DEMO_febio_0063_custom_hip_implant_01.m
Below is a demonstration for:
- Importing bone geometry
- Building geometry for a custom hip implant
- Evaluating the implant using FEA
Contents
- Keywords
- Plot settings
- Control parameters
- Import bone model
- Compute derived control parameters
- Cut bone end off
- Cut femoral head off
- Add inner bone surface for implant to rest on (will attach to collar)
- Create implant spherical head
- Build colar
- Build neck
- Define shaft
- Join bone and implant surfaces
- Create tetrahedral elements for both regions
- Define material regions in bone
- Visualizing solid mesh
- Load SED data from non-implant model and access SED interpolation function
- Find femoral head
- Work out force distribution on femoral head surface nodes
- Marking muscle locations
- Muscle force definition
- Defining prescribed forces
- Visualizing boundary conditions
- Defining the FEBio input structure
- Quick viewing of the FEBio input file structure
- Exporting the FEBio input file
- Running the FEBio analysis
- Import FEBio results
- Compare to non-implant case
- simulated results using anim8 to visualize and animate
Keywords
- febio_spec version 4.0
- febio, FEBio
- beam force loading
- force control boundary condition
- tetrahedral elements, tet4
- static, solid
- hyperelastic, Ogden
- displacement logfile
- stress logfile
clear; close all; clc;
Plot settings
fontSize=20; faceAlpha1=0.8; markerSize=40; markerSize2=20; lineWidth=3;
Control parameters
% Path names defaultFolder = fileparts(fileparts(mfilename('fullpath'))); savePath=fullfile(defaultFolder,'data','temp'); pathNameSTL=fullfile(defaultFolder,'data','STL'); loadName_SED=fullfile(savePath,'SED_no_implant.mat'); if ~exist(loadName_SED,'file') skipCompare=1; warning('This demo requires: DEMO_febio_0062_femur_load_01.m, to be completed first. Demo will continue but comparison to hip without femur will be skipped.'); else skipCompare=0; end % Defining file names febioFebFileNamePart='tempModel'; febioFebFileName=fullfile(savePath,[febioFebFileNamePart,'.feb']); %FEB file name febioLogFileName=fullfile(savePath,[febioFebFileNamePart,'.txt']); %FEBio log file name febioLogFileName_disp=[febioFebFileNamePart,'_disp_out.txt']; %Log file name for exporting displacancellousBone febioLogFileName_force=[febioFebFileNamePart,'_force_out.txt']; %Log file name for exporting force febioLogFileName_strainEnergy=[febioFebFileNamePart,'_energy_out.txt']; %Log file name for exporting strain energy density febioLogFileName_stress=[febioFebFileNamePart,'_stress_out.txt']; %Log file name for exporting stress %Geometric parameters distanceCut=250; %Distance from femur to cut bone at corticalThickness=3; %Thickness used for cortical material definition volumeFactors=[2 2]; %Factor to scale desired volume for interior elements w.r.t. boundary elements stemCut=32; numSmoothStepsCutBottom=5; numSmoothStepsCutFemur=25; implantOffset=5; implantOffsetCollar=2; implantHeadRadius=20; implantStickRadius=10; implantCollarThickness=3; implantShaftLengthFraction=1/4; numLoftGuideSlicesShaft=8; numSmoothStepsShaft=20; %Define applied force forceTotal=[-405 -246 -1717.5]; %x,y,z force in Newton forceAbductor=[564.831 -132.696 704.511]; forceVastusLateralis_Walking=[-7.857 -161.505 -811.017]; forceVastusLateralis_StairClimbing=[-19.206 -195.552 -1,179.423]; forceVastusMedialis_StairClimbing=[-76.824 -345.708 -2,331.783]; forceVM_inactive=[0 0 0]; n=1; switch n case 1 forceVastusLateralis=forceVastusLateralis_Walking; forceVastusMedialis=forceVM_inactive; case 2 forceVastusLateralis=forceVastusLateralis_StairClimbing; forceVastusMedialis=forceVastusMedialis_StairClimbing; otherwise forceVastusLateralis=forceVastusLateralis_StairClimbing; forceVastusMedialis=forceVastusMedialis_StairClimbing; end % Distance markers and scaling factor zLevelWidthMeasure = -75; zLevelFCML = -395; scaleFactorSize=1; distanceMuscleAttachAbductor=15; distanceMuscleVastusLateralis=10; distanceMuscleAttachVastusMedialis=10; %Material parameters (MPa if spatial units are mm) % Cortical bone E_youngs1=17000; %Youngs modulus nu1=0.25; %Poissons ratio % Cancellous bone E_youngs2=1500; %Youngs modulus nu2=0.25; %Poissons ratio % Implant E_youngs3=110000; %Youngs modulus nu3=0.25; %Poissons ratio % FEA control settings numTimeSteps=10; %Number of time steps desired max_refs=25; %Max reforms max_ups=0; %Set to zero to use full-Newton iterations opt_iter=6; %Optimum number of iterations max_retries=5; %Maximum number of retires dtmin=(1/numTimeSteps)/100; %Minimum time step size dtmax=1/numTimeSteps; %Maximum time step size runMode='external'; %'external' or 'internal'
Import bone model
[stlStruct] = import_STL(fullfile(pathNameSTL,'femur_iso.stl')); F_bone=stlStruct.solidFaces{1}; %Faces V_bone=stlStruct.solidVertices{1}; %Vertices V_bone=V_bone.*1000; [F_bone,V_bone]=mergeVertices(F_bone,V_bone); % Merging nodes Q1=euler2DCM([0 0 0.065*pi]); V_bone=V_bone*Q1; Q2=euler2DCM([-0.5*pi 0 0]); V_bone=V_bone*Q2; Q3=euler2DCM([0 0 0.36*pi]); V_bone=V_bone*Q3;
Compute derived control parameters
pointSpacing=mean(patchEdgeLengths(F_bone,V_bone)); %Points spacing of bone surface snapTolerance=mean(patchEdgeLengths(F_bone,V_bone))/100; %Tolerance for surface slicing femurHeight=max(V_bone(:,3))-min(V_bone(:,3)); %Height of the femur shaftDepthFromHead=femurHeight.*implantShaftLengthFraction;
Cut bone end off
%Cut bone [F_bone,V_bone,~,logicSide,~]=triSurfSlice(F_bone,V_bone,[],[0 0 -distanceCut],[0 0 1]); F_bone=F_bone(logicSide==0,:); [F_bone,V_bone]=patchCleanUnused(F_bone,V_bone); %Get boundary curve Eb=patchBoundary(F_bone); indCurve=edgeListToCurve(Eb); indCurve=indCurve(1:end-1); %Smooth curve edges cparSmooth.n=numSmoothStepsCutBottom; cparSmooth.Method='HC'; [V_Eb_smooth]=patchSmooth(Eb,V_bone(:,[1 2]),[],cparSmooth); V_bone(indCurve,[1 2])=V_Eb_smooth(indCurve,:); %Smooth mesh at curve clear cparSmooth cparSmooth.n=numSmoothStepsCutBottom; cparSmooth.Method='HC'; cparSmooth.RigidConstraints=indCurve; [V_bone]=patchSmooth(F_bone,V_bone,[],cparSmooth); %Mesh the bottom curfaces [F_bone2,V_bone2]=regionTriMesh3D({V_bone(indCurve,:)},pointSpacing,0,'linear'); if dot(mean(patchNormal(F_bone2,V_bone2)),[0 0 1])>0 F_bone2=fliplr(F_bone2); end %Join cut bone with bottom to created a single node shared closed surface [F_bone,V_bone,C_bone]=joinElementSets({F_bone,F_bone2},{V_bone,V_bone2}); [F_bone,V_bone]=mergeVertices(F_bone,V_bone);
Visualize cut surface
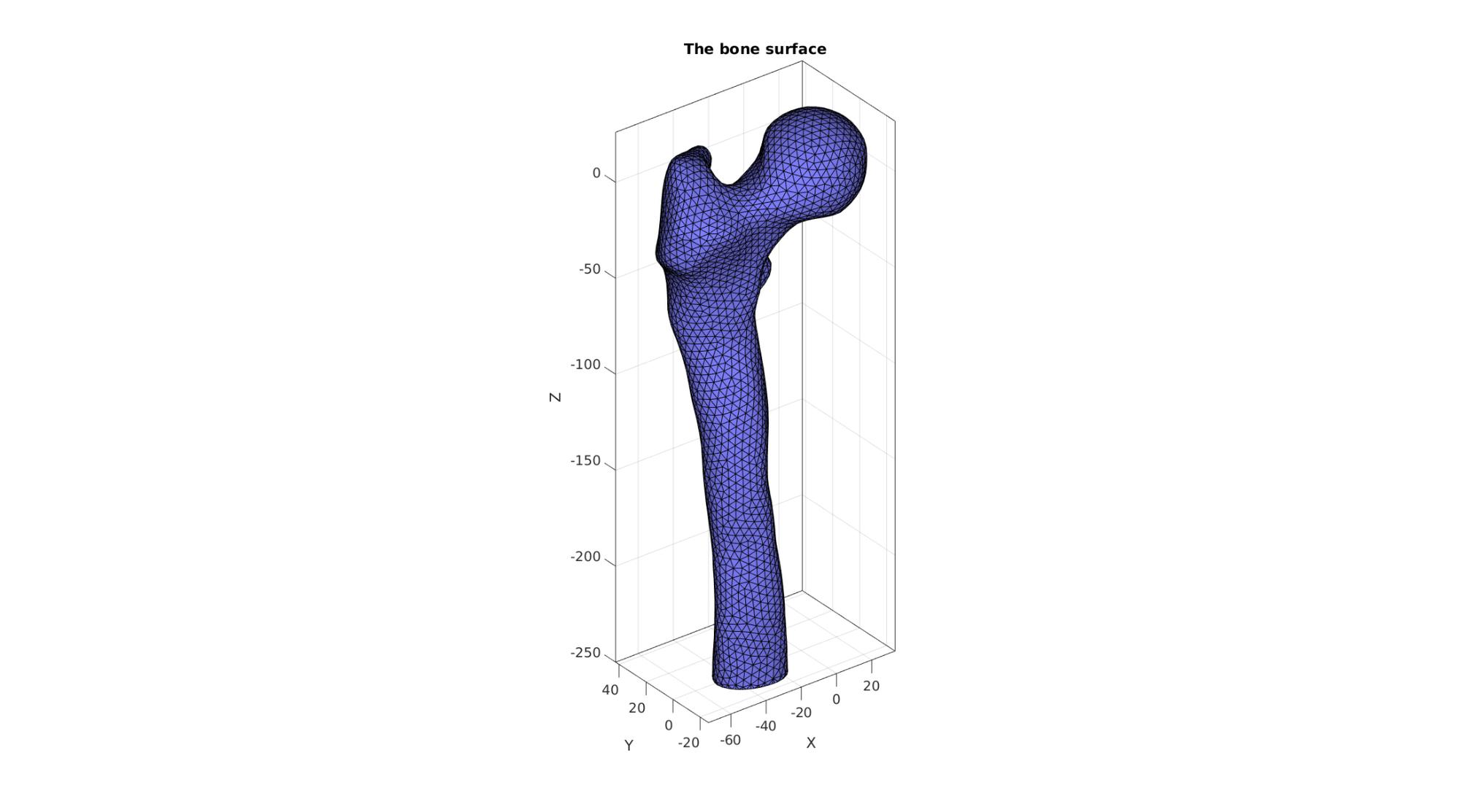
cFigure; hold on; title('The bone surface'); gpatch(F_bone,V_bone,'bw','k',1); axisGeom; camlight headlight; gdrawnow;
Cut femoral head off
%Set-up orientation and location of cutting plane n=vecnormalize([0 0 1]); %Normal direction to plane Q1=euler2DCM([0 -(63/180)*pi 0]); Q2=euler2DCM([0 0 (32/180)*pi]); Q=Q1*Q2; n=n*Q; P_cut=[0 0 0]-n*stemCut; %Point on plane %Slicing surface [Fc,Vc,Cc,logicSide,~]=triSurfSlice(F_bone,V_bone,C_bone,P_cut,n,snapTolerance); %Compose isolated cut geometry and boundary curves [F_bone_cut,V_bone_cut]=patchCleanUnused(Fc(logicSide,:),Vc); C_bone_cut=Cc(logicSide); Eb=patchBoundary(F_bone_cut); indCutCurve=edgeListToCurve(Eb); indCutCurve=indCutCurve(1:end-1); %Smoothing cut logicTouch=any(ismember(F_bone_cut,indCutCurve),2); indTouch=unique(F_bone_cut(logicTouch,:)); logicTouch=any(ismember(F_bone_cut,indTouch),2); indRigid=unique(F_bone_cut(~logicTouch,:)); indRigid=unique([indRigid(:);indCutCurve(:)]); clear cparSmooth cparSmooth.n=numSmoothStepsCutFemur; cparSmooth.RigidConstraints=indRigid; [V_bone_cut]=patchSmooth(F_bone_cut,V_bone_cut,[],cparSmooth); clear cparSmooth cparSmooth.n=numSmoothStepsCutFemur; [VEb]=patchSmooth(Eb,V_bone_cut,[],cparSmooth); V_bone_cut(indCutCurve,:)=VEb(indCutCurve,:); %Get curve points and centre coordinate V_bone_cut_curve=V_bone_cut(indCutCurve,:); PA=mean(V_bone_cut_curve,1); %Mean of cut curve (middle of cut)
Visualize cut surface
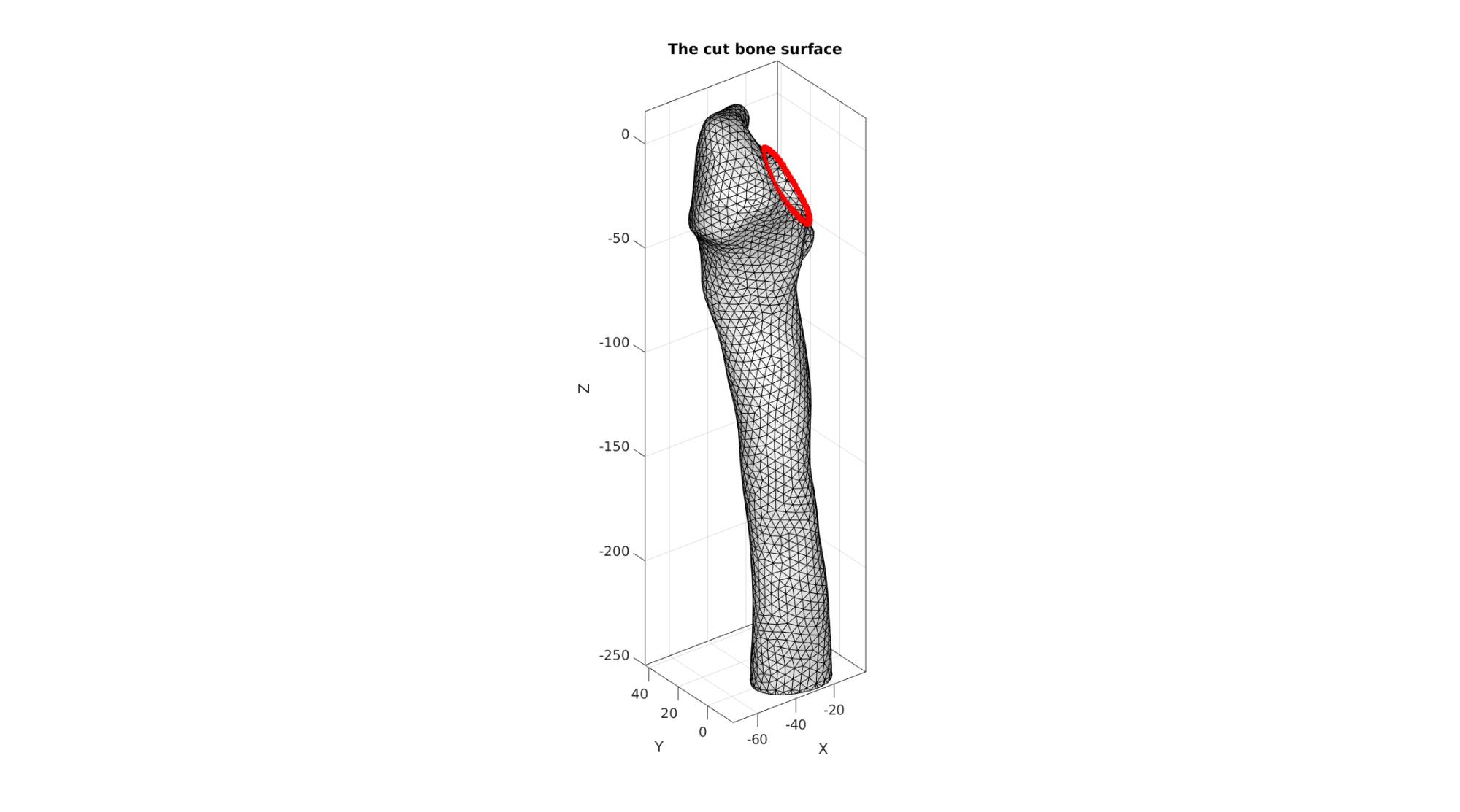
cFigure; hold on; title('The cut bone surface'); gpatch(F_bone_cut,V_bone_cut,'w','k',1); plotV(V_bone_cut_curve,'r.-','MarkerSize',25,'LineWidth',3) axisGeom; camlight headlight; gdrawnow;
Add inner bone surface for implant to rest on (will attach to collar)
%Define collar curve V_collar_1=V_bone_cut_curve; V_collar_1=V_collar_1-PA; d=sqrt(sum(V_collar_1.^2,2)); shrinkFactor=(min(d(:))-implantOffsetCollar)./min(d(:)); V_collar_1=V_collar_1.*shrinkFactor; V_collar_1=V_collar_1+PA; V_collar_1=evenlySpaceCurve(V_collar_1,pointSpacing,'pchip',1); % Mesh cut bone top (space between collar curve and bone cut curve) [F_bone_top,V_bone_top]=regionTriMesh3D({V_bone_cut_curve,V_collar_1},[],0,'linear'); nTop=mean(patchNormal(F_bone_top,V_bone_top),1); if dot(nTop,n)<0 F_bone_top=fliplr(F_bone_top); end % Join and merge geometry [F_bone_cut,V_bone_cut,C_bone_cut]=joinElementSets({F_bone_cut,F_bone_top},{V_bone_cut,V_bone_top},{C_bone_cut,max(C_bone_cut(:))+ones(size(F_bone_top,1),1)}); [F_bone_cut,V_bone_cut]=mergeVertices(F_bone_cut,V_bone_cut);
Visualize bone with meshed top
cFigure; hold on; title('The cut bone surface with meshed top'); gpatch(F_bone_cut,V_bone_cut,C_bone_cut,'k',1); plotV(V_bone_cut_curve,'r.-','MarkerSize',15,'LineWidth',2) plotV(V_collar_1,'g.-','MarkerSize',15,'LineWidth',2) axisGeom; icolorbar; camlight headlight; gdrawnow;

Create implant spherical head
% Create sphere use will loop to define increasingly fine sphere % triangulations untill the point spacing of the sphere is smaller or equal % to the bone point spacing. nRef=1; while 1 [F_head,V_head]=geoSphere(nRef,implantHeadRadius); pointSpacingNow=mean(patchEdgeLengths(F_head,V_head)); if pointSpacingNow<=pointSpacing break else nRef=nRef+1; end end
Cutting the sphere
% Define ball cutting coordinate xc=cos(asin(implantStickRadius/implantHeadRadius))*implantHeadRadius; logicRight=V_head(:,1)>xc; logicCut=any(logicRight(F_head),2); logicCut=triSurfLogicSharpFix(F_head,logicCut,3); F_head=F_head(~logicCut,:); [F_head,V_head]=patchCleanUnused(F_head,V_head); Eb_head=patchBoundary(F_head); indB=unique(Eb_head(:)); [T,P,R] = cart2sph(V_head(:,2),V_head(:,3),V_head(:,1)); P(indB)=atan2(xc,implantHeadRadius*sin(acos(xc./implantHeadRadius))); [V_head(:,2),V_head(:,3),V_head(:,1)] = sph2cart(T,P,R); indHeadHole=edgeListToCurve(Eb_head); indHeadHole=indHeadHole(1:end-1); pointSpacingHead=mean(sqrt(sum(diff(V_head(indHeadHole,:),1,1).^2,2))); %Reorient ball nd=vecnormalize(-PA); %Vector pointing to mean of cut curve Q=vecPair2Rot(nd,[0 0 1]); %Rotation matrix for ball Q3=euler2DCM([0 -0.5*pi 0]); V_head=V_head*Q3; V_head=V_head*Q;
Visualize head
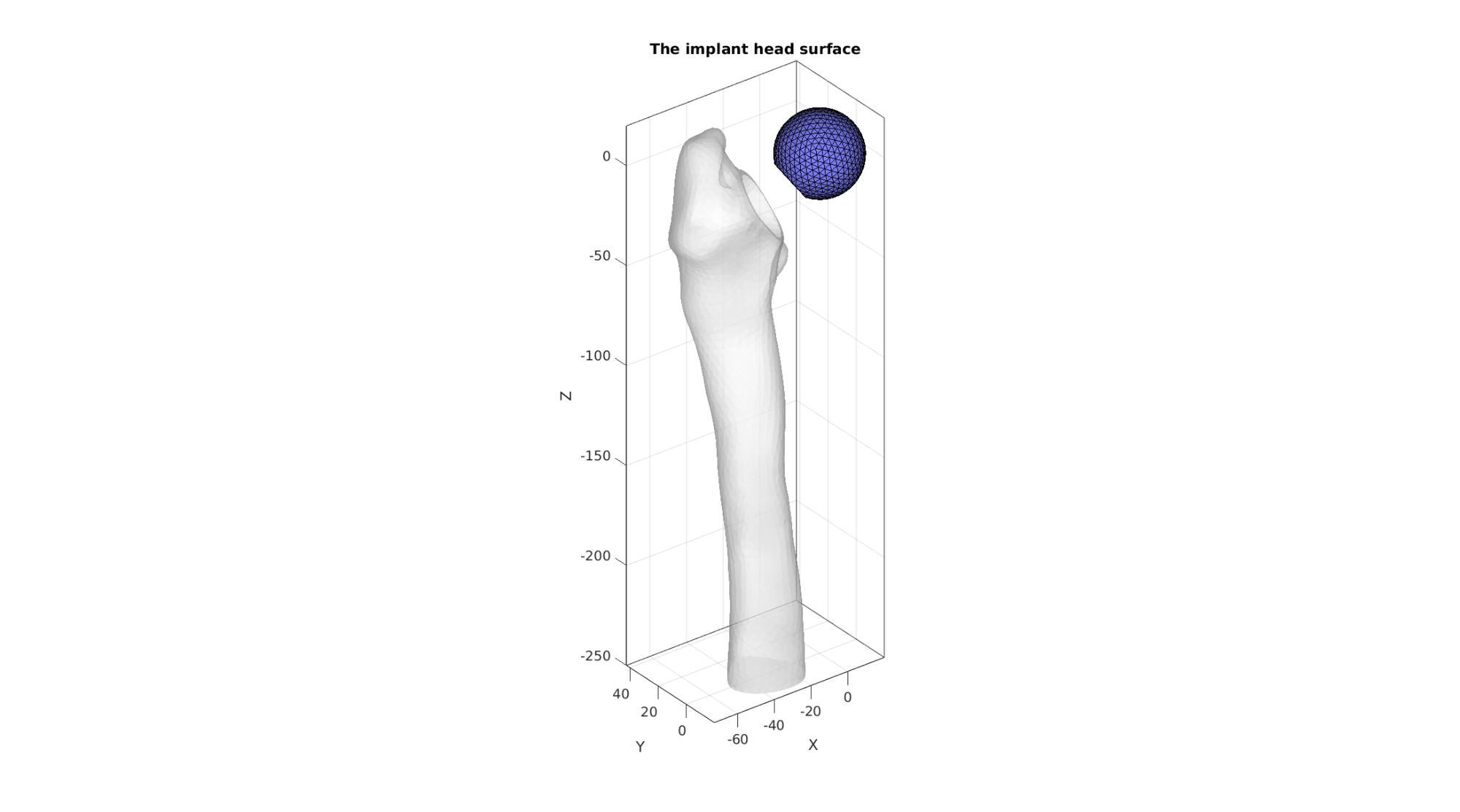
cFigure; hold on; title('The implant head surface'); gpatch(F_bone_cut,V_bone_cut,'w','none',0.5); gpatch(F_head,V_head,'bw','k',1); axisGeom; camlight headlight; gdrawnow;
Build colar
%Create color offset curve V_colarTop=V_collar_1; ve=[V_colarTop(2:end,:); V_colarTop(1,:)]-V_colarTop; vd=vecnormalize(cross(n(ones(size(ve,1),1),:),ve)); % vn2(:,1)=vn2(:,1)+collarThickness.*n; V_colarTop=V_colarTop+implantCollarThickness.*n; nc=ceil((pi*implantCollarThickness/2)/pointSpacingHead); if nc<4 nc=4; end vc=linspacen(V_collar_1,V_colarTop,nc); X=squeeze(vc(:,1,:)); Y=squeeze(vc(:,2,:)); Z=squeeze(vc(:,3,:)); t=repmat(linspace(-1,1,nc),size(Z,1),1); a=acos(t); X=X+implantCollarThickness/2.*sin(a).*repmat(vd(:,1),1,nc); Y=Y+implantCollarThickness/2.*sin(a).*repmat(vd(:,2),1,nc); Z=Z+implantCollarThickness/2.*sin(a).*repmat(vd(:,3),1,nc); [F_collar,V_collar]=grid2patch(X,Y,Z,[],[1 0 0]); [F_collar,V_collar]=quad2tri(F_collar,V_collar,'a'); indTopCollar=size(V_collar,1)+1-size(V_collar_1,1):size(V_collar,1); Eb_collar=[indTopCollar(:) [indTopCollar(2:end)'; indTopCollar(1)]];
Visualize collar
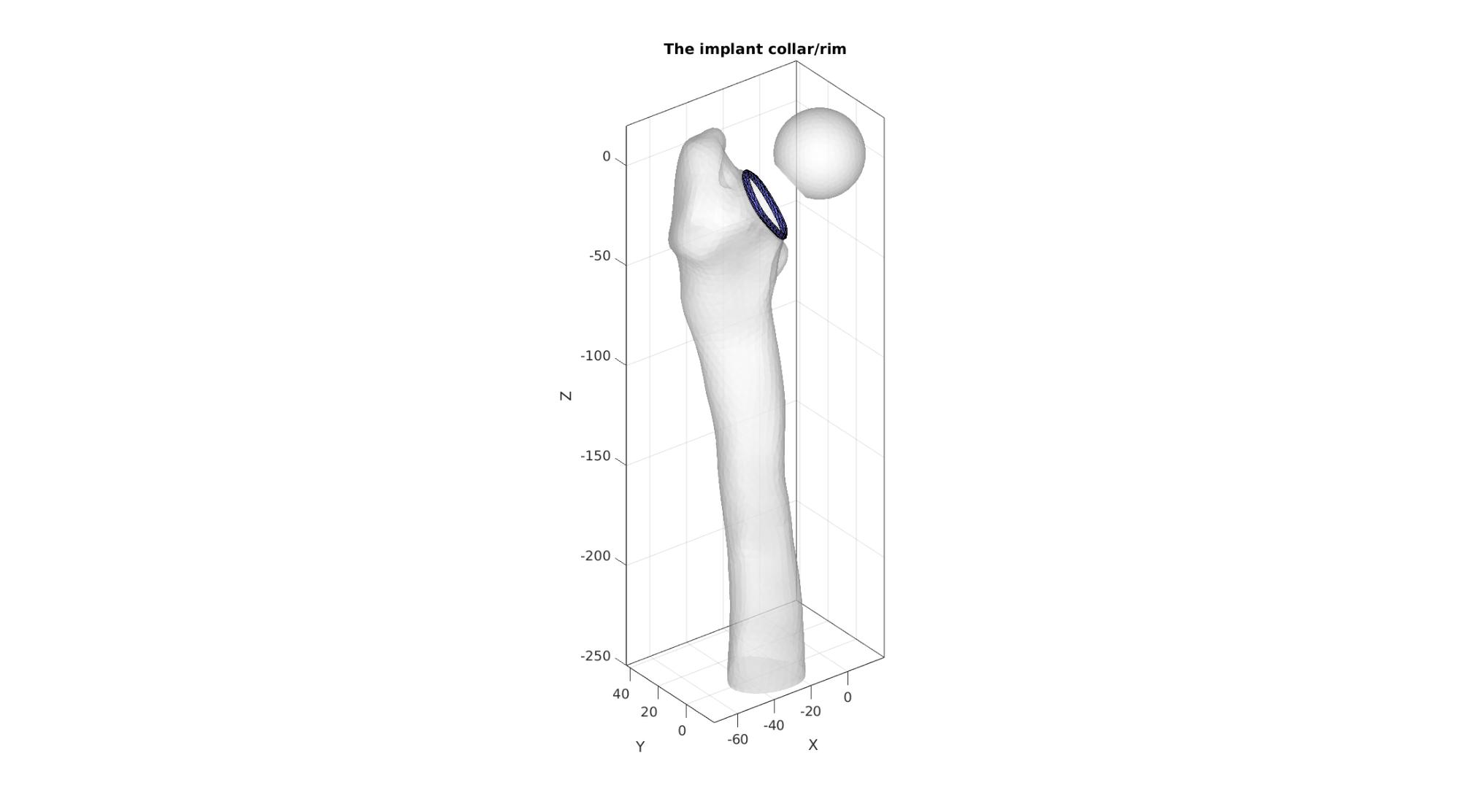
cFigure; hold on; title('The implant collar/rim'); gpatch(F_bone_cut,V_bone_cut,'w','none',0.5); gpatch(F_head,V_head,'w','none',0.5); gpatch(F_collar,V_collar,'bw','k',1); axisGeom; camlight headlight; gdrawnow;
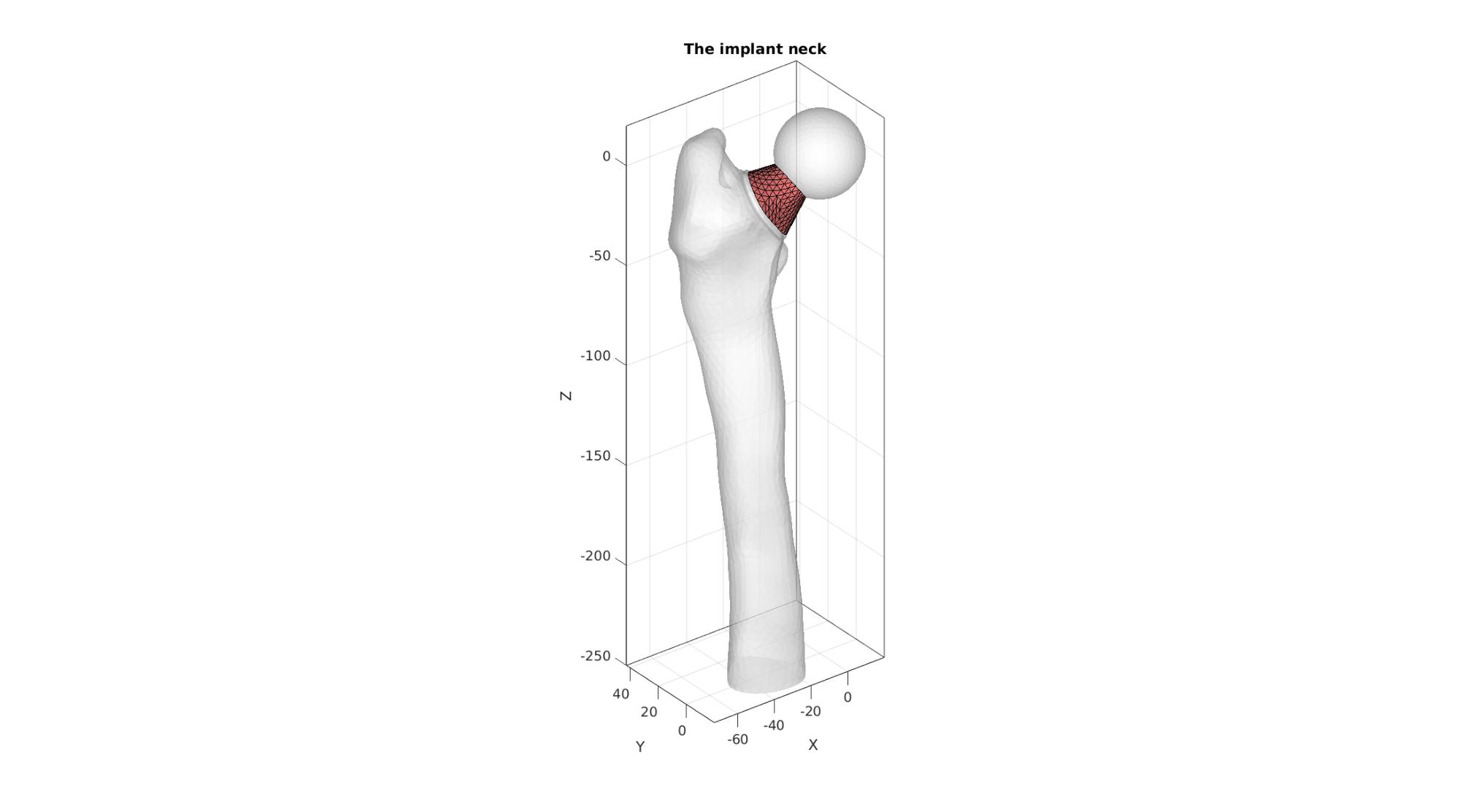
Build neck
if size(Eb_head,1)<size(Eb_collar,1) numEdgesNeeded=size(Eb_collar,1); [F_head,V_head,Eb_head,~]=triSurfSplitBoundary(F_head,V_head,Eb_head,numEdgesNeeded,[]); elseif size(Eb_collar,1)<size(Eb_head,1) numEdgesNeeded=size(Eb_head,1); [F_collar,V_collar,Eb_collar,~]=triSurfSplitBoundary(F_collar,V_collar,Eb_collar,numEdgesNeeded,[]); end indTopCollar=edgeListToCurve(Eb_collar); indTopCollar=indTopCollar(1:end-1); indHeadHole=edgeListToCurve(Eb_head); indHeadHole=indHeadHole(1:end-1); [~,indStart]=minDist(V_collar(indTopCollar(1),:),V_head(indHeadHole,:)); if indStart>1 indHeadHole=[indHeadHole(indStart:end) indHeadHole(1:indStart-1)]; end d1=sum((V_head(indHeadHole,:)-V_collar(indTopCollar,:)).^2,2); d2=sum((V_head(indHeadHole,:)-V_collar(flip(indTopCollar),:)).^2,2); if max(d2(:))<max(d1(:)) indTopCollar=flip(indTopCollar); end cParLoft.closeLoopOpt=1; cParLoft.patchType='tri_slash'; [F_neck,V_neck,indNeckStart,indNeckEnd]=polyLoftLinear(V_collar(indTopCollar,:),V_head(indHeadHole,:),cParLoft);
Visualize collar
cFigure; hold on; title('The implant neck'); gpatch(F_bone_cut,V_bone_cut,'w','none',0.5); gpatch(F_head,V_head,'w','none',0.5); gpatch(F_collar,V_collar,'w','none',0.5); gpatch(F_neck,V_neck,'rw','k',1); axisGeom; camlight headlight; gdrawnow;
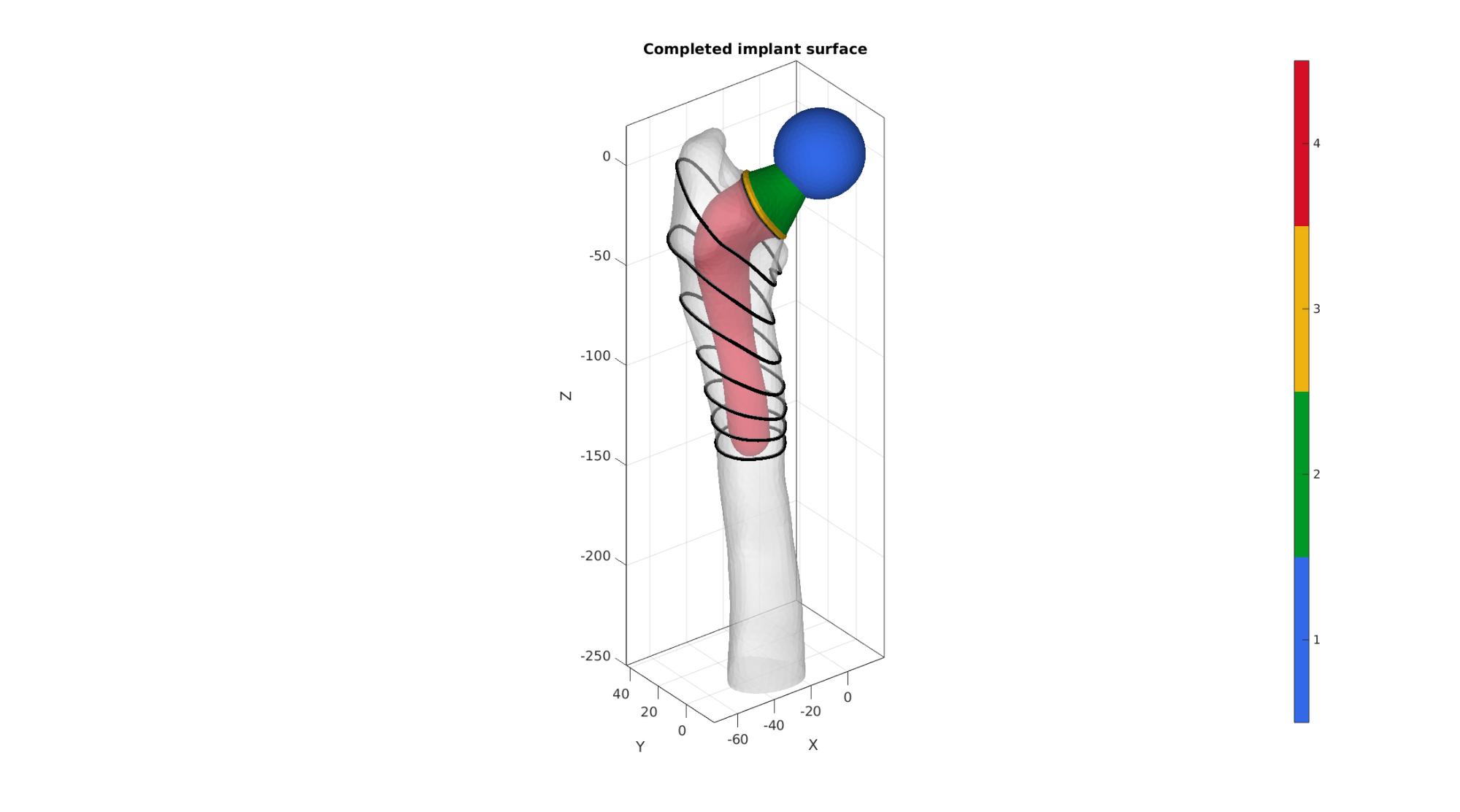
Define shaft
%Find lowest point on collar curve [~,indMin]=min(V_collar_1(:,3)); PB=V_collar_1(indMin,:); %Define a normalized vector pointing from PA to PB m=vecnormalize(PB-PA); %Define based point for slicing f=shaftDepthFromHead./abs(m(3)); P=PA+f*m; %Define normal directions for slicing (go linearly from n to z-dir) nn=linspacen(n,[0 0 1],numLoftGuideSlicesShaft)'; numPointsRadial=size(V_collar_1,1); Vp=V_collar_1; FL=[]; VL=Vp; [~,indStart]=max(VL(:,1)); if indStart>1 VL=[VL(indStart:end,:); VL(1:indStart-1,:)]; end R1=vecPair2Rot(n,[0 0 1]); e=[(1:numPointsRadial)' [(2:numPointsRadial)';1]]; V_start=VL(1,:); t=linspace(0,2*pi,numPointsRadial+1); t=t(1:end-1); VC=cell(numLoftGuideSlicesShaft,1); VC{1}=V_collar_1; for q=2:1:numLoftGuideSlicesShaft Rn=vecPair2Rot(nn(q,:),[0 0 1]); [~,Vqc,~,~,Ebc]=triSurfSlice(F_bone,V_bone,[],P,nn(q,:),snapTolerance); indCutCurveNow=edgeListToCurve(Ebc); indCutCurveNow=indCutCurveNow(1:end-1); P2=mean(Vqc(indCutCurveNow,:),1); if P2(3)>-50 Vp=V_collar_1-PA; Vp=Vp*R1'; Vp=Vp*Rn; else Vp=Vqc(indCutCurveNow,:); Vp=Vp-P2; d=sqrt(sum(Vp.^2,2)); shrinkFactor=(min(d(:))-implantOffset)./min(d(:)); Vp=Vp.*shrinkFactor; end Vp=Vp+P2; Vp=evenlySampleCurve(Vp,numPointsRadial,0.01,1); [~,indStart]=max(Vp(:,1)); if indStart>1 Vp=[Vp(indStart:end,:); Vp(1:indStart-1,:)]; end f=[e+(q-2)*numPointsRadial fliplr(e)+(q-1)*numPointsRadial]; FL=[FL;f]; VL=[VL;Vp]; V_start=Vp(1,:); VC{q}=Vqc(indCutCurveNow,:); end [Fe,Ve]=regionTriMesh3D({Vp},[],0,'linear'); ne=mean(patchNormal(Fe,Ve),1); if dot(ne,[0 0 1])>0 Fe=fliplr(Fe); end d=minDist(Ve,Vp); r=max(d(:)); d=d./max(d(:)); a=acos(1-d); Ve(:,3)=Ve(:,3)-r*sin(a); [FL,VL]=subQuad(FL,VL,3,3); [FL,VL]=quad2tri(FL,VL,'a'); [FL,VL]=joinElementSets({FL,Fe},{VL,Ve}); [FL,VL]=mergeVertices(FL,VL); Eb=patchBoundary(FL); cparSmooth.n=numSmoothStepsShaft; % cPar.Method='HC'; cparSmooth.RigidConstraints=unique(Eb(:)); VL=patchSmooth(FL,VL,[],cparSmooth); % Join and merge surfaces [F_implant,V_implant,C_implant]=joinElementSets({F_head,F_neck,F_collar,FL},{V_head,V_neck,V_collar,VL}); [F_implant,V_implant]=mergeVertices(F_implant,V_implant);
Visualize implant
cFigure; hold on; title('Completed implant surface'); gpatch(F_bone_cut,V_bone_cut,'w','none',0.5); gpatch(F_implant,V_implant,C_implant,'none',1); for q=1:1:numLoftGuideSlicesShaft Vpp=VC{q}; plotV(Vpp([1:size(Vpp,1) 1],:),'k-','LineWidth',4); end axisGeom; camlight headlight; colormap(gjet(250)); icolorbar; gdrawnow;
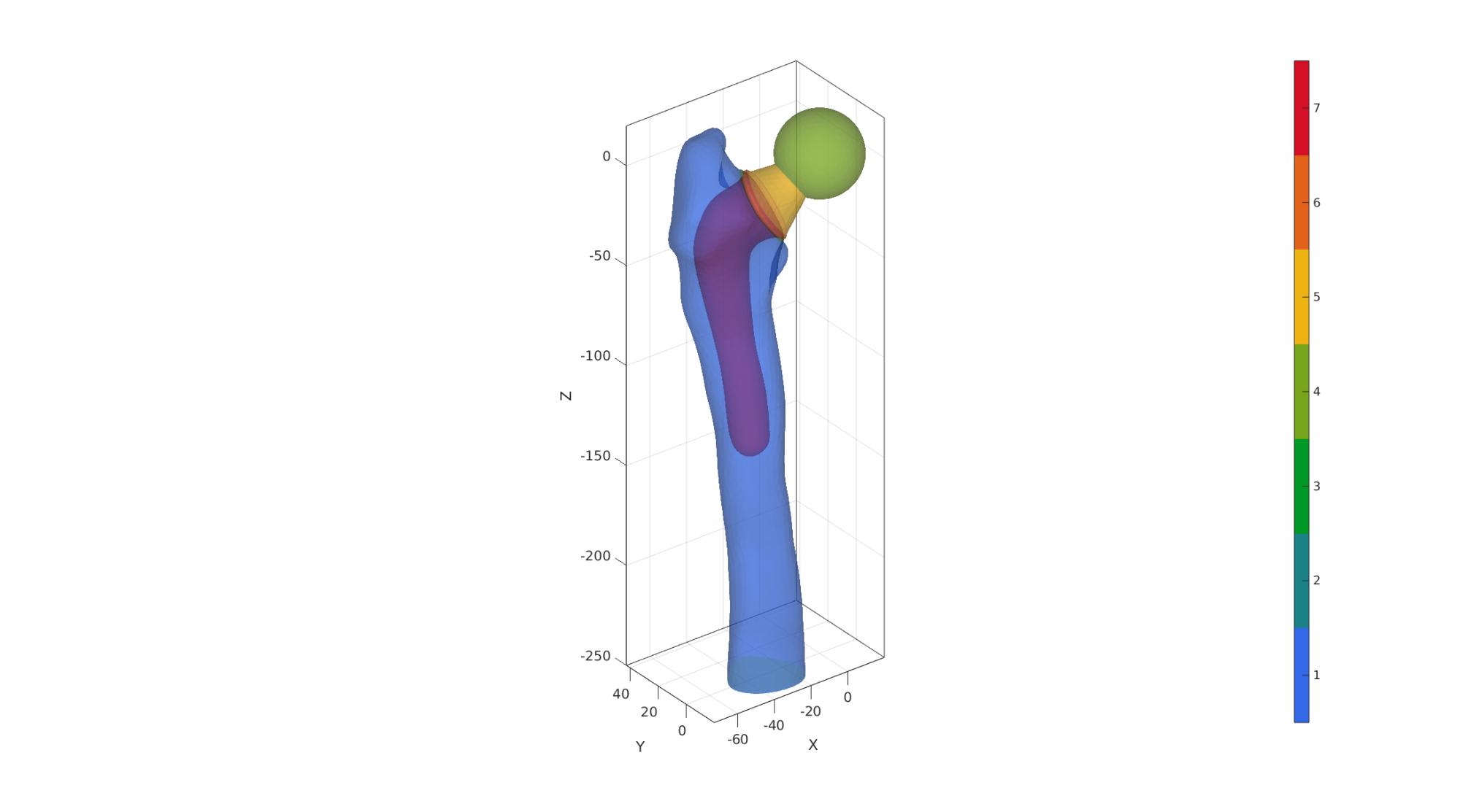
Join bone and implant surfaces
[F,V,C]=joinElementSets({F_bone_cut,F_implant},{V_bone_cut,V_implant},{C_bone_cut,C_implant+max(C_bone_cut(:))});
[F,V]=mergeVertices(F,V);
Visualized merged set
cFigure; hold on; gpatch(F,V,C,'none',0.5); axisGeom; camlight headlight; colormap(gjet(250)); icolorbar; gdrawnow;
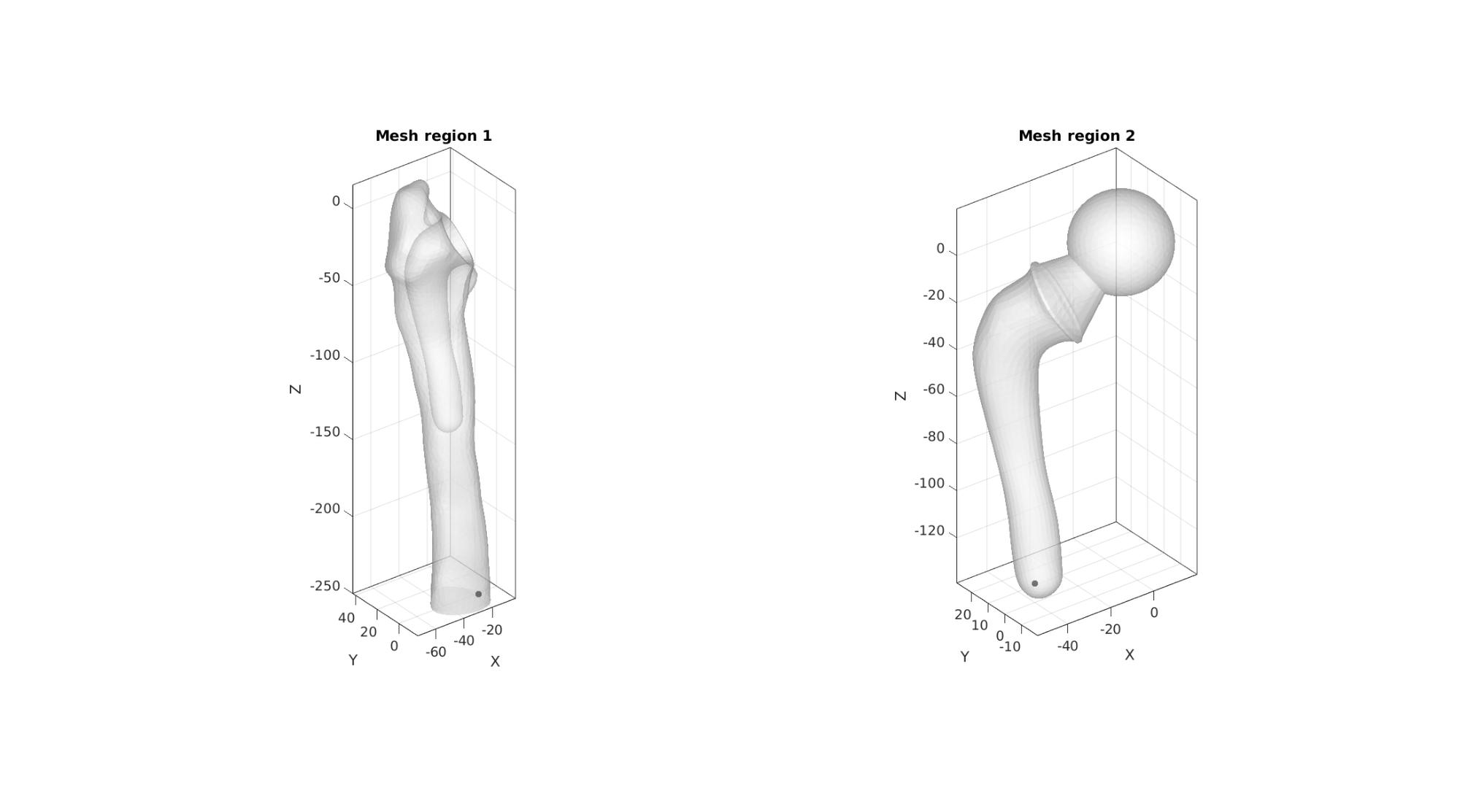
Create tetrahedral elements for both regions
Define interior points
logicRegion1=ismember(C,[1 2 3 7]); %Bone region is defined by bone+shaft surfaces V_region1=getInnerPoint(F(logicRegion1,:),V); logicRegion2=ismember(C,4:7); %Logic for implant faces V_region2=getInnerPoint(F(logicRegion2,:),V); V_regions=[V_region1; V_region2];
Visualize mesh regions and interior points
cFigure; subplot(1,2,1); hold on; title('Mesh region 1'); gpatch(F(logicRegion1,:),V,'w','none',0.5); plotV(V_region1,'k.','MarkerSize',25); axisGeom; camlight headlight; subplot(1,2,2); hold on; title('Mesh region 2'); gpatch(F(logicRegion2,:),V,'w','none',0.5); plotV(V_region2,'k.','MarkerSize',25); axisGeom; camlight headlight; gdrawnow;
Mesh using TetGen
regionTetVolumes=volumeFactors.*tetVolMeanEst(F,V); %Create tetgen input structure inputStruct.stringOpt='-pq1.2AaY'; %Options for tetgen inputStruct.Faces=F; %Boundary faces inputStruct.Nodes=V; %Nodes of boundary inputStruct.faceBoundaryMarker=C; inputStruct.regionPoints=V_regions; %Interior points for regions inputStruct.holePoints=[]; %Interior points for holes inputStruct.regionA=regionTetVolumes; %Desired tetrahedral volume for each region % Mesh model using tetrahedral elements using tetGen [meshOutput]=runTetGen(inputStruct); %Run tetGen
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- TETGEN Tetrahedral meshing --- 29-May-2023 10:39:08 %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- Writing SMESH file --- 29-May-2023 10:39:08 ----> Adding node field ----> Adding facet field ----> Adding holes specification ----> Adding region specification --- Done --- 29-May-2023 10:39:08 --- Running TetGen to mesh input boundary--- 29-May-2023 10:39:08 Opening /home/kevin/DATA/Code/matlab/GIBBON/data/temp/temp.smesh. Delaunizing vertices... Delaunay seconds: 0.020498 Creating surface mesh ... Surface mesh seconds: 0.004431 Recovering boundaries... Boundary recovery seconds: 0.008259 Removing exterior tetrahedra ... Spreading region attributes. Exterior tets removal seconds: 0.003486 Recovering Delaunayness... Delaunay recovery seconds: 0.006234 Refining mesh... 6679 insertions, added 5728 points, 197840 tetrahedra in queue. 2224 insertions, added 1564 points, 211485 tetrahedra in queue. 2964 insertions, added 1320 points, 85537 tetrahedra in queue. Refinement seconds: 0.131803 Smoothing vertices... Mesh smoothing seconds: 0.223018 Improving mesh... Mesh improvement seconds: 0.008581 Writing /home/kevin/DATA/Code/matlab/GIBBON/data/temp/temp.1.node. Writing /home/kevin/DATA/Code/matlab/GIBBON/data/temp/temp.1.ele. Writing /home/kevin/DATA/Code/matlab/GIBBON/data/temp/temp.1.face. Writing /home/kevin/DATA/Code/matlab/GIBBON/data/temp/temp.1.edge. Output seconds: 0.032528 Total running seconds: 0.439064 Statistics: Input points: 5011 Input facets: 10046 Input segments: 15054 Input holes: 0 Input regions: 2 Mesh points: 13953 Mesh tetrahedra: 75470 Mesh faces: 154199 Mesh faces on exterior boundary: 6518 Mesh faces on input facets: 10046 Mesh edges on input segments: 15054 Steiner points inside domain: 8942 --- Done --- 29-May-2023 10:39:09 %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- Importing TetGen files --- 29-May-2023 10:39:09 --- Done --- 29-May-2023 10:39:09
Access mesh output structure
E=meshOutput.elements; %The elements V=meshOutput.nodes; %The vertices or nodes CE=meshOutput.elementMaterialID; %Element material or region id Fb=meshOutput.facesBoundary; %The boundary faces Cb=meshOutput.boundaryMarker; %The boundary markers
Define material regions in bone
logicImplantElements=CE==-3; indBoundary=unique(Fb(Cb==1,:)); DE=minDist(V,V(indBoundary,:)); logicCorticalNodes=DE<=corticalThickness; logicCorticalElements=any(logicCorticalNodes(E),2) & ~logicImplantElements; logicCancellousElements=~logicCorticalElements & ~logicImplantElements; E1=E(logicCorticalElements,:); E2=E(logicCancellousElements,:); E3=E(logicImplantElements,:); E=[E1;E2;E3]; elementMaterialID=[ones(size(E1,1),1);2*ones(size(E2,1),1);3*ones(size(E3,1),1)]; logicBoneElements=ismember(elementMaterialID,[1 2]); E_12=E(logicBoneElements,:); %Only bone elements VE_12=patchCentre(E_12,V); %Centre coordinates for bone elements meshOutput.elements=E; meshOutput.elementMaterialID=elementMaterialID;
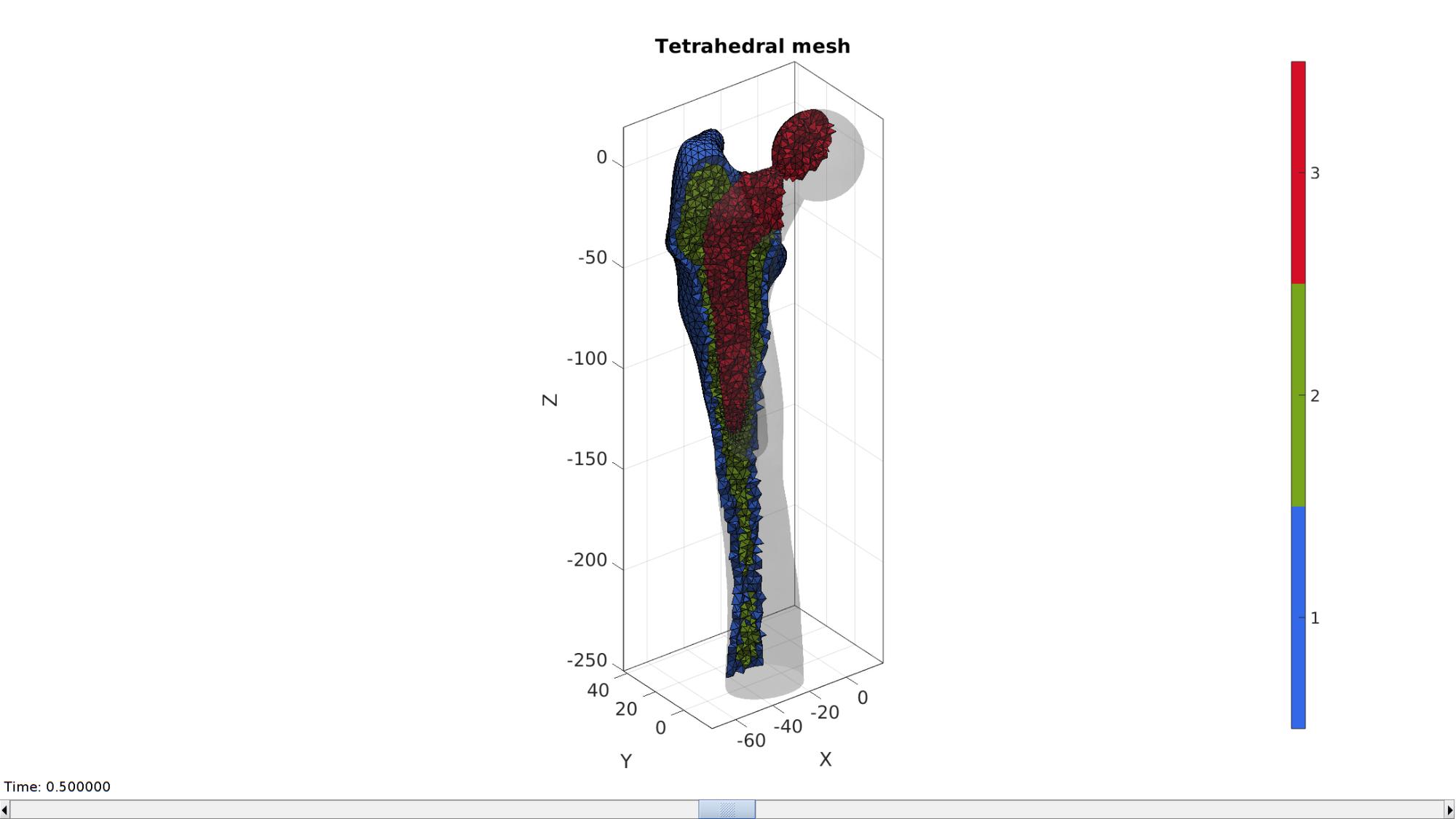
Visualizing solid mesh
hFig=cFigure; hold on;
optionStruct.hFig=hFig;
meshView(meshOutput,optionStruct);
axisGeom;
drawnow;

Load SED data from non-implant model and access SED interpolation function
if skipCompare==0 %Load data without implant outputStruct=load(loadName_SED); Fb_no_implant = outputStruct.boundaryFaces; V_no_implant = outputStruct.nodes; interpFuncEnergy=outputStruct.interpFuncEnergy; % Interpolation function end
Visualize solid mesh
hf=cFigure; hold on; title('Tetrahedral mesh','FontSize',fontSize); % Visualizing using |meshView| optionStruct.hFig=hf; meshView(meshOutput,optionStruct); axisGeom(gca,fontSize); gdrawnow;
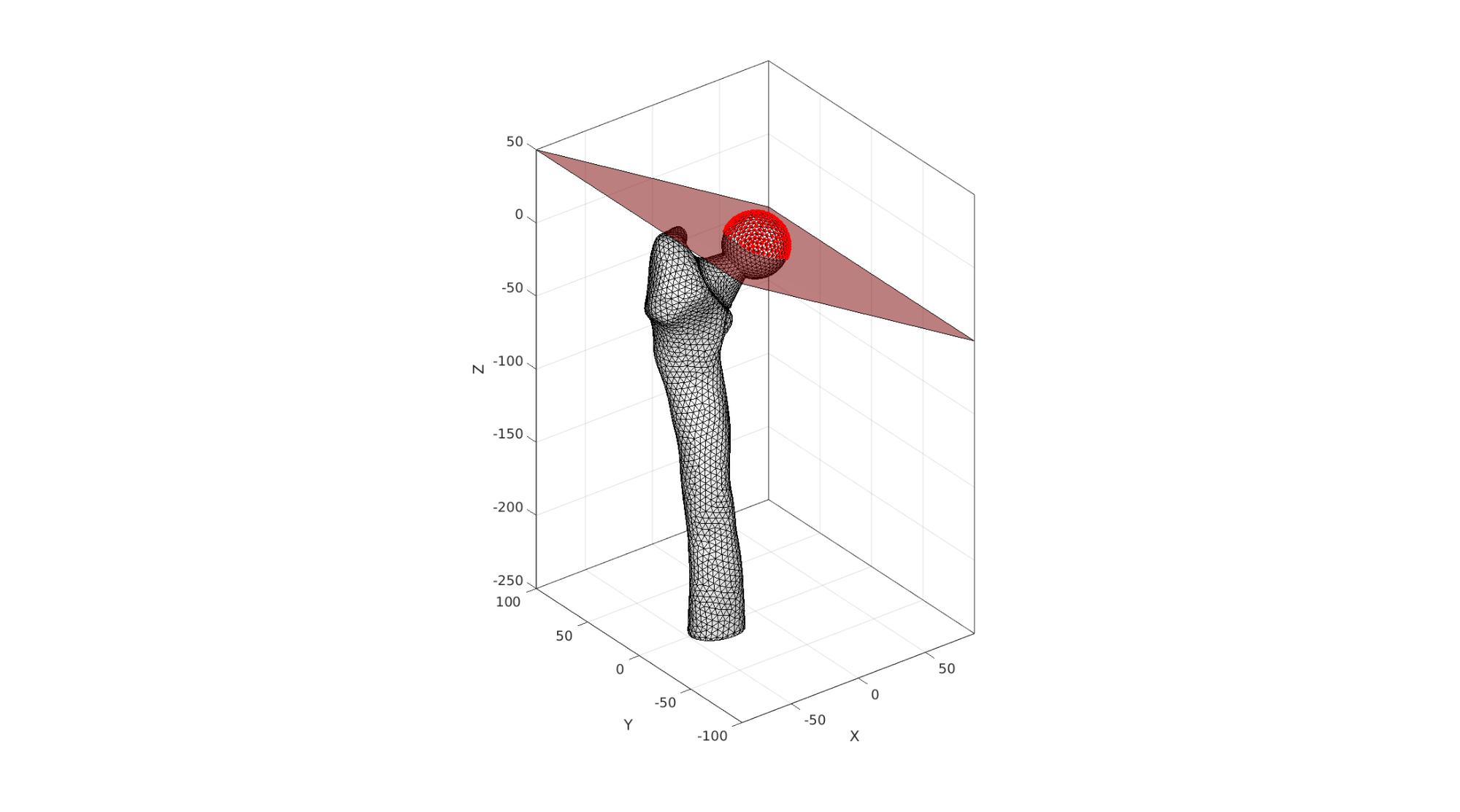
Find femoral head
w=100; f=[1 2 3 4]; v=w*[-1 -1 0; -1 1 0; 1 1 0; 1 -1 0]; p=[0 0 0]; Q=euler2DCM([0 (150/180)*pi 0]); v=v*Q; v=v+p; Vr=V*Q'; Vr=Vr+p; logicHeadNodes=Vr(:,3)<0; logicHeadFaces=all(logicHeadNodes(Fb),2); bcPrescribeList_head=unique(Fb(logicHeadFaces,:));
Visualize femoral head nodes for prescribed force boundary conditions
cFigure; hold on; gpatch(Fb,V,'w','k',1); gpatch(f,v,'r','k',0.5); plotV(V(bcPrescribeList_head,:),'r.','markerSize',15) axisGeom; camlight headlight; drawnow;
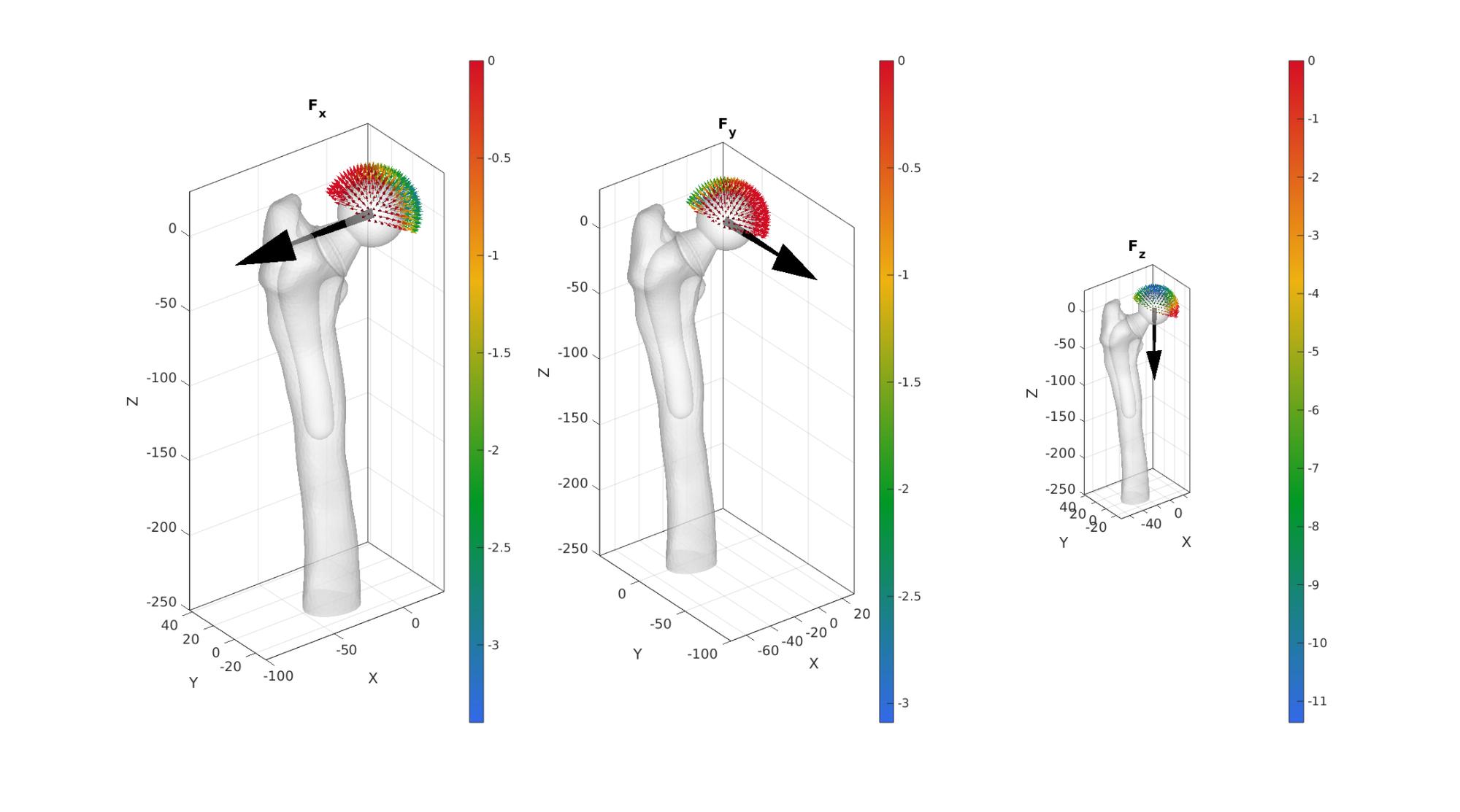
Work out force distribution on femoral head surface nodes
This is based on surface normal directions. Forces are assumed to only be able to act in a compressive sense on the bone.
[~,~,N]=patchNormal(fliplr(Fb),V); %Nodal normal directions FX=[forceTotal(1) 0 0]; %X force vector FY=[0 forceTotal(2) 0]; %Y force vector FZ=[0 0 forceTotal(3)]; %Z force vector wx=dot(N(bcPrescribeList_head,:),FX(ones(numel(bcPrescribeList_head),1),:),2); wy=dot(N(bcPrescribeList_head,:),FY(ones(numel(bcPrescribeList_head),1),:),2); wz=dot(N(bcPrescribeList_head,:),FZ(ones(numel(bcPrescribeList_head),1),:),2); %Force zero wx(wx>0)=0; wy(wy>0)=0; wz(wz>0)=0; force_X=forceTotal(1).*ones(numel(bcPrescribeList_head),1).*wx; force_Y=forceTotal(2).*ones(numel(bcPrescribeList_head),1).*wy; force_Z=forceTotal(3).*ones(numel(bcPrescribeList_head),1).*wz; force_X=force_X./sum(force_X(:)); %sum now equal to 1 force_X=force_X.*forceTotal(1); %sum now equal to desired force_Y=force_Y./sum(force_Y(:)); %sum now equal to 1 force_Y=force_Y.*forceTotal(2); %sum now equal to desired force_Z=force_Z./sum(force_Z(:)); %sum now equal to 1 force_Z=force_Z.*forceTotal(3); %sum now equal to desired force_head=[force_X(:) force_Y(:) force_Z(:)];
cFigure; subplot(1,3,1);hold on; title('F_x'); gpatch(Fb,V,'w','none',0.5); quiverVec([0 0 0],FX,100,'k'); % scatterV(V(indicesHeadNodes,:),15) quiverVec(V(bcPrescribeList_head,:),N(bcPrescribeList_head,:),10,force_X); axisGeom; camlight headlight; colormap(gca,gjet(250)); colorbar; subplot(1,3,2);hold on; title('F_y'); gpatch(Fb,V,'w','none',0.5); quiverVec([0 0 0],FY,100,'k'); % scatterV(V(indicesHeadNodes,:),15) quiverVec(V(bcPrescribeList_head,:),N(bcPrescribeList_head,:),10,force_Y); axisGeom; camlight headlight; colormap(gca,gjet(250)); colorbar; subplot(1,3,3);hold on; title('F_z'); gpatch(Fb,V,'w','none',0.5); quiverVec([0 0 0],FZ,100,'k'); % scatterV(V(indicesHeadNodes,:),15) quiverVec(V(bcPrescribeList_head,:),N(bcPrescribeList_head,:),10,force_Z); axisGeom; camlight headlight; colormap(gca,gjet(250)); colorbar; drawnow;
Marking muscle locations
P_abductor_find = [-69.771045288206111 8.185179717034659 -5.575329878303917]; %Coordinate at centre of muscle attachment [~,indAbductor]=minDist(P_abductor_find,V); %Node number of point at centre of attachment dAbductor=meshDistMarch(Fb,V,indAbductor); %Distance (on mesh) from attachement centre bcPrescibeList_abductor=find(dAbductor<=distanceMuscleAttachAbductor); %Node numbers for attachment site P_VastusLateralis_find = [-58.763839901506827 19.145444610053566 -51.005278396808819]; %Coordinate at centre of muscle attachment [~,indVastusLateralis]=minDist(P_VastusLateralis_find,V); %Node number of point at centre of attachment dVastusLateralis=meshDistMarch(Fb,V,indVastusLateralis); %Distance (on mesh) from attachement centre bcPrescibeList_VastusLateralis=find(dVastusLateralis<=distanceMuscleVastusLateralis); %Node numbers for attachment site P_VastusMedialis_find = [-18.533631492778085 9.501312355952791 -85.666499329588035]; %Coordinate at centre of muscle attachment [~,indVastusMedialis]=minDist(P_VastusMedialis_find,V); %Node number of point at centre of attachment dVastusMedialis=meshDistMarch(Fb,V,indVastusMedialis); %Distance (on mesh) from attachement centre bcPrescibeList_VastusMedialis=find(dVastusMedialis<=distanceMuscleAttachVastusMedialis); %Node numbers for attachment site
Muscle force definition
forceAbductor_distributed=forceAbductor.*ones(numel(bcPrescibeList_abductor),1)./numel(bcPrescibeList_abductor); forceVastusLateralis_distributed=forceVastusLateralis.*ones(numel(bcPrescibeList_VastusLateralis),1)./numel(bcPrescibeList_VastusLateralis); forceVastusMedialis_distributed=forceVastusMedialis.*ones(numel(bcPrescibeList_VastusMedialis),1)./numel(bcPrescibeList_VastusMedialis);
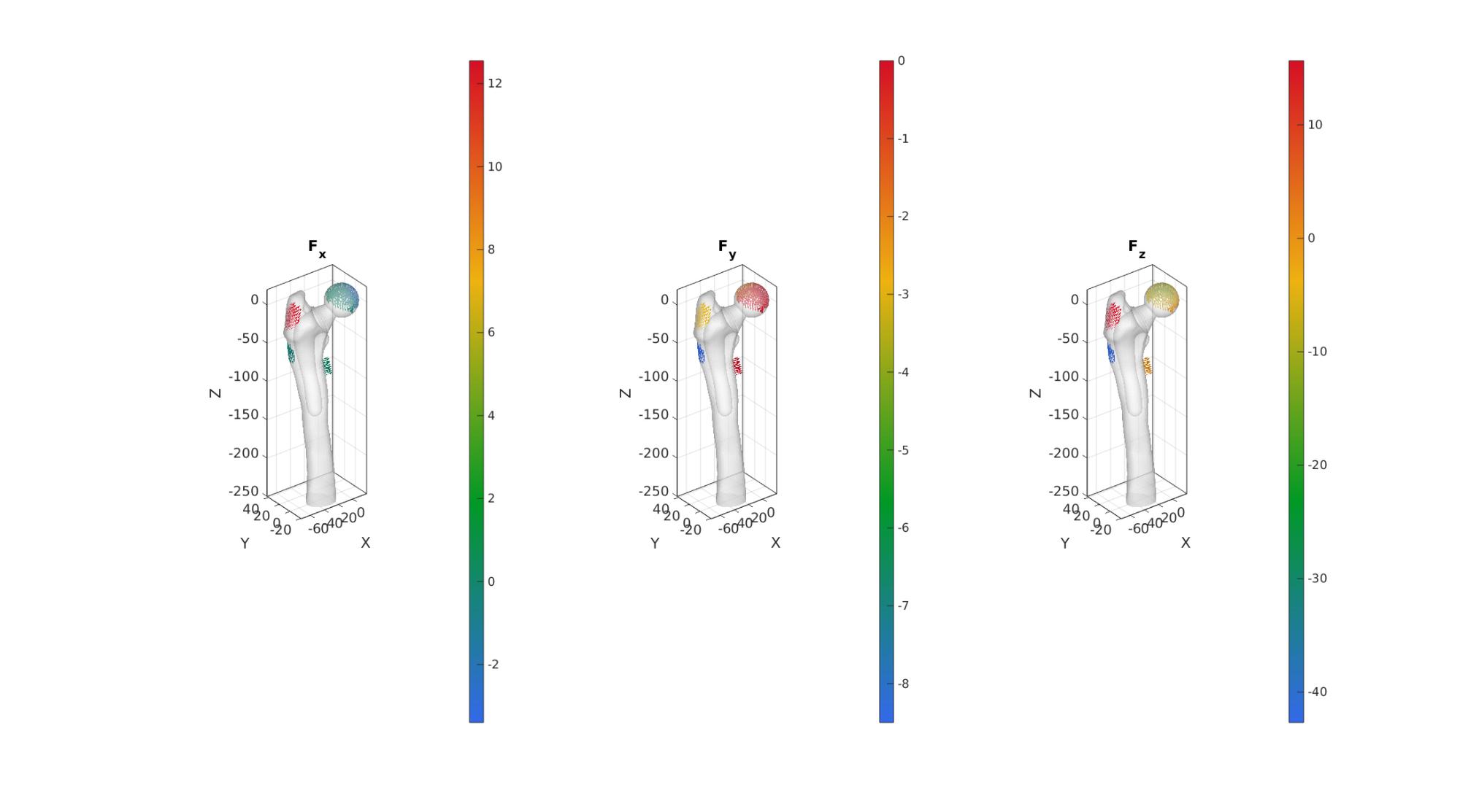
Defining prescribed forces
bcPrescribeList=[bcPrescribeList_head(:);... bcPrescibeList_abductor(:);... bcPrescibeList_VastusLateralis(:);... bcPrescibeList_VastusMedialis(:)]; forceData=[force_head;... forceAbductor_distributed;... forceVastusLateralis_distributed;... forceVastusMedialis_distributed];
cFigure; subplot(1,3,1);hold on; title('F_x'); gpatch(Fb,V,'w','none',0.5); quiverVec(V(bcPrescribeList,:),forceData,10,forceData(:,1)); axisGeom; camlight headlight; colormap(gca,gjet(250)); colorbar; subplot(1,3,2);hold on; title('F_y'); gpatch(Fb,V,'w','none',0.5); quiverVec(V(bcPrescribeList,:),forceData,10,forceData(:,2)); axisGeom; camlight headlight; colormap(gca,gjet(250)); colorbar; subplot(1,3,3);hold on; title('F_z'); gpatch(Fb,V,'w','none',0.5); quiverVec(V(bcPrescribeList,:),forceData,10,forceData(:,3)); axisGeom; camlight headlight; colormap(gca,gjet(250)); colorbar;
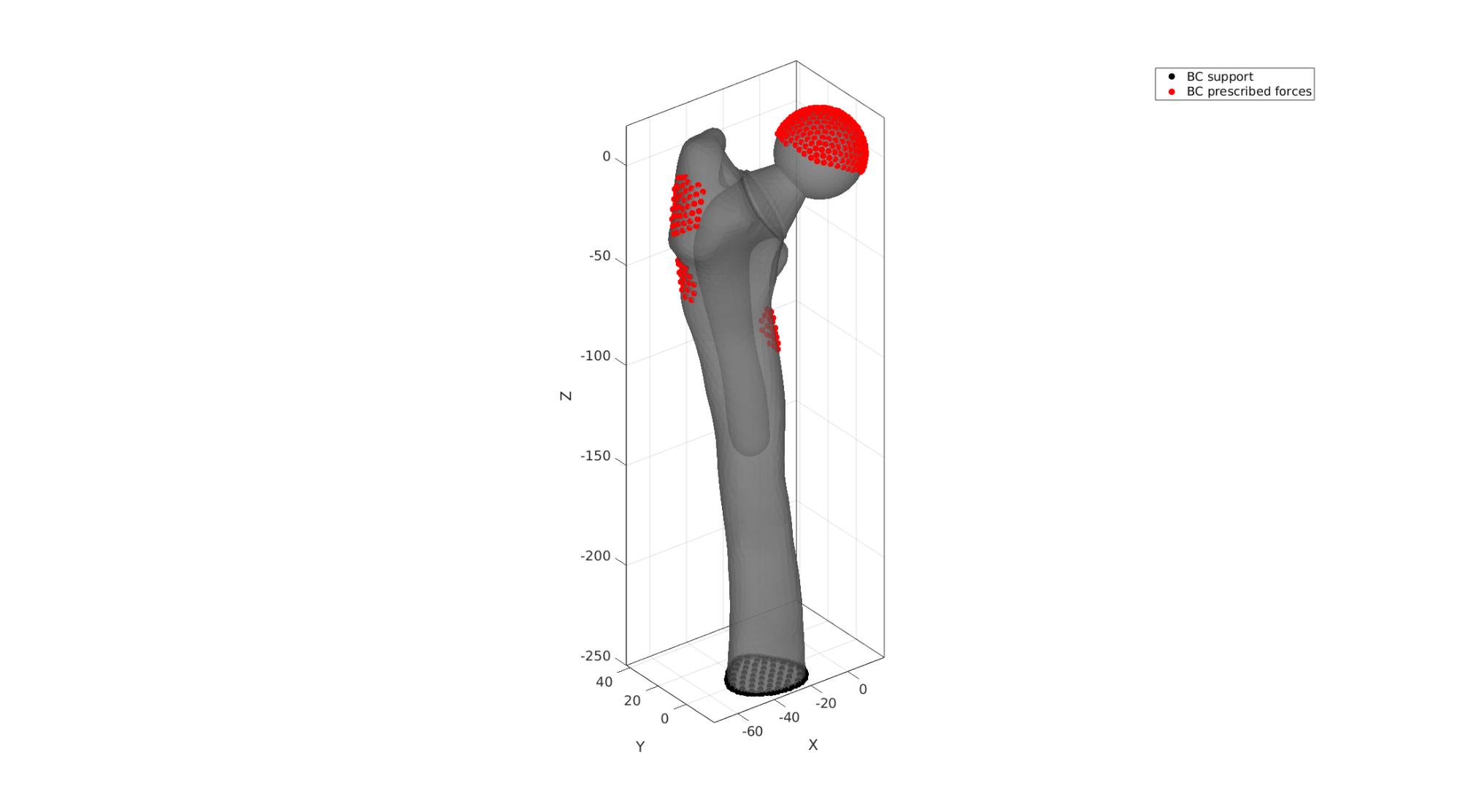
Visualizing boundary conditions
F_bottomSupport=Fb(Cb==2,:); bcSupportList=unique(F_bottomSupport(:)); cFigure; hold on; gpatch(Fb,V,'kw','none',0.7); hl(1)=plotV(V(bcSupportList,:),'k.','MarkerSize',25); hl(2)=plotV(V(bcPrescribeList,:),'r.','MarkerSize',25); legend(hl,{'BC support','BC prescribed forces'}); axisGeom; camlight headlight; drawnow;
Defining the FEBio input structure
See also febioStructTemplate and febioStruct2xml and the FEBio user manual.
%Get a template with default settings [febio_spec]=febioStructTemplate; %febio_spec version febio_spec.ATTR.version='4.0'; %Module section febio_spec.Module.ATTR.type='solid'; %Control section febio_spec.Control.analysis='STATIC'; febio_spec.Control.time_steps=numTimeSteps; febio_spec.Control.step_size=1/numTimeSteps; febio_spec.Control.solver.max_refs=max_refs; febio_spec.Control.solver.qn_method.max_ups=max_ups; febio_spec.Control.time_stepper.dtmin=dtmin; febio_spec.Control.time_stepper.dtmax=dtmax; febio_spec.Control.time_stepper.max_retries=max_retries; febio_spec.Control.time_stepper.opt_iter=opt_iter; %Material section materialName1='Material1'; febio_spec.Material.material{1}.ATTR.name=materialName1; febio_spec.Material.material{1}.ATTR.type='neo-Hookean'; febio_spec.Material.material{1}.ATTR.id=1; febio_spec.Material.material{1}.E=E_youngs1; febio_spec.Material.material{1}.v=nu1; materialName2='Material2'; febio_spec.Material.material{2}.ATTR.name=materialName2; febio_spec.Material.material{2}.ATTR.type='neo-Hookean'; febio_spec.Material.material{2}.ATTR.id=2; febio_spec.Material.material{2}.E=E_youngs2; febio_spec.Material.material{2}.v=nu2; materialName3='Material3'; febio_spec.Material.material{3}.ATTR.name=materialName3; febio_spec.Material.material{3}.ATTR.type='neo-Hookean'; febio_spec.Material.material{3}.ATTR.id=3; febio_spec.Material.material{3}.E=E_youngs3; febio_spec.Material.material{3}.v=nu3; % Mesh section % -> Nodes febio_spec.Mesh.Nodes{1}.ATTR.name='Object1'; %The node set name febio_spec.Mesh.Nodes{1}.node.ATTR.id=(1:size(V,1))'; %The node id's febio_spec.Mesh.Nodes{1}.node.VAL=V; %The nodel coordinates % -> Elements partName1='CorticalBone'; febio_spec.Mesh.Elements{1}.ATTR.name=partName1; %Name of this part febio_spec.Mesh.Elements{1}.ATTR.type='tet4'; %Element type febio_spec.Mesh.Elements{1}.elem.ATTR.id=(1:1:size(E1,1))'; %Element id's febio_spec.Mesh.Elements{1}.elem.VAL=E1; %The element matrix partName2='CancellousBone'; febio_spec.Mesh.Elements{2}.ATTR.name=partName2; %Name of this part febio_spec.Mesh.Elements{2}.ATTR.type='tet4'; %Element type febio_spec.Mesh.Elements{2}.elem.ATTR.id=size(E1,1)+(1:1:size(E2,1))'; %Element id's febio_spec.Mesh.Elements{2}.elem.VAL=E2; %The element matrix partName3='Implant'; febio_spec.Mesh.Elements{3}.ATTR.name=partName3; %Name of this part febio_spec.Mesh.Elements{3}.ATTR.type='tet4'; %Element type febio_spec.Mesh.Elements{3}.elem.ATTR.id=size(E1,1)+size(E2,1)+(1:1:size(E3,1))'; %Element id's febio_spec.Mesh.Elements{3}.elem.VAL=E3; %The element matrix % -> NodeSets nodeSetName1='bcSupportList'; febio_spec.Mesh.NodeSet{1}.ATTR.name=nodeSetName1; febio_spec.Mesh.NodeSet{1}.VAL=mrow(bcSupportList); nodeSetName2='bcPrescribeList'; febio_spec.Mesh.NodeSet{2}.ATTR.name=nodeSetName2; febio_spec.Mesh.NodeSet{2}.VAL=mrow(bcPrescribeList); %MeshDomains section febio_spec.MeshDomains.SolidDomain{1}.ATTR.name=partName1; febio_spec.MeshDomains.SolidDomain{1}.ATTR.mat=materialName1; febio_spec.MeshDomains.SolidDomain{2}.ATTR.name=partName2; febio_spec.MeshDomains.SolidDomain{2}.ATTR.mat=materialName2; febio_spec.MeshDomains.SolidDomain{3}.ATTR.name=partName3; febio_spec.MeshDomains.SolidDomain{3}.ATTR.mat=materialName3; %Boundary condition section % -> Fix boundary conditions febio_spec.Boundary.bc{1}.ATTR.name='zero_displacement_xyz'; febio_spec.Boundary.bc{1}.ATTR.type='zero displacement'; febio_spec.Boundary.bc{1}.ATTR.node_set=nodeSetName1; febio_spec.Boundary.bc{1}.x_dof=1; febio_spec.Boundary.bc{1}.y_dof=1; febio_spec.Boundary.bc{1}.z_dof=1; %MeshData secion %-> Node data loadDataName1='nodal_forces'; febio_spec.MeshData.NodeData{1}.ATTR.name=loadDataName1; febio_spec.MeshData.NodeData{1}.ATTR.node_set=nodeSetName2; febio_spec.MeshData.NodeData{1}.ATTR.data_type='vec3'; febio_spec.MeshData.NodeData{1}.node.ATTR.lid=(1:1:numel(bcPrescribeList))'; febio_spec.MeshData.NodeData{1}.node.VAL=forceData; %Loads section % -> Prescribed nodal forces febio_spec.Loads.nodal_load{1}.ATTR.name='PrescribedForce'; febio_spec.Loads.nodal_load{1}.ATTR.type='nodal_force'; febio_spec.Loads.nodal_load{1}.ATTR.node_set=nodeSetName2; febio_spec.Loads.nodal_load{1}.value.ATTR.lc=1; febio_spec.Loads.nodal_load{1}.value.ATTR.type='map'; febio_spec.Loads.nodal_load{1}.value.VAL=loadDataName1; %LoadData section % -> load_controller febio_spec.LoadData.load_controller{1}.ATTR.name='LC_1'; febio_spec.LoadData.load_controller{1}.ATTR.id=1; febio_spec.LoadData.load_controller{1}.ATTR.type='loadcurve'; febio_spec.LoadData.load_controller{1}.interpolate='LINEAR'; %febio_spec.LoadData.load_controller{1}.extend='CONSTANT'; febio_spec.LoadData.load_controller{1}.points.pt.VAL=[0 0; 1 1]; %Output section % -> log file febio_spec.Output.logfile.ATTR.file=febioLogFileName; febio_spec.Output.logfile.node_data{1}.ATTR.file=febioLogFileName_disp; febio_spec.Output.logfile.node_data{1}.ATTR.data='ux;uy;uz'; febio_spec.Output.logfile.node_data{1}.ATTR.delim=','; febio_spec.Output.logfile.element_data{1}.ATTR.file=febioLogFileName_stress; febio_spec.Output.logfile.element_data{1}.ATTR.data='s1;s2;s3'; febio_spec.Output.logfile.element_data{1}.ATTR.delim=','; febio_spec.Output.logfile.element_data{2}.ATTR.file=febioLogFileName_strainEnergy; febio_spec.Output.logfile.element_data{2}.ATTR.data='sed'; febio_spec.Output.logfile.element_data{2}.ATTR.delim=','; % Plotfile section febio_spec.Output.plotfile.compression=0;
Quick viewing of the FEBio input file structure
The febView function can be used to view the xml structure in a MATLAB figure window.
febView(febio_spec); %Viewing the febio file
Exporting the FEBio input file
Exporting the febio_spec structure to an FEBio input file is done using the febioStruct2xml function.
febioStruct2xml(febio_spec,febioFebFileName); %Exporting to file and domNode
Running the FEBio analysis
To run the analysis defined by the created FEBio input file the runMonitorFEBio function is used. The input for this function is a structure defining job settings e.g. the FEBio input file name. The optional output runFlag informs the user if the analysis was run succesfully.
febioAnalysis.run_filename=febioFebFileName; %The input file name febioAnalysis.run_logname=febioLogFileName; %The name for the log file febioAnalysis.disp_on=1; %Display information on the command window febioAnalysis.runMode=runMode; [runFlag]=runMonitorFEBio(febioAnalysis);%START FEBio NOW!!!!!!!!
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
--------> RUNNING/MONITORING FEBIO JOB <-------- 29-May-2023 10:39:18
FEBio path: /home/kevin/FEBioStudio/bin/febio4
# Attempt removal of existing log files 29-May-2023 10:39:18
* Removal succesful 29-May-2023 10:39:18
# Attempt removal of existing .xplt files 29-May-2023 10:39:18
* Removal succesful 29-May-2023 10:39:18
# Starting FEBio... 29-May-2023 10:39:18
Max. total analysis time is: Inf s
* Waiting for log file creation 29-May-2023 10:39:18
Max. wait time: 30 s
* Log file found. 29-May-2023 10:39:19
# Parsing log file... 29-May-2023 10:39:19
number of iterations : 3 29-May-2023 10:39:22
number of reformations : 3 29-May-2023 10:39:22
------- converged at time : 0.1 29-May-2023 10:39:22
number of iterations : 3 29-May-2023 10:39:22
number of reformations : 3 29-May-2023 10:39:22
------- converged at time : 0.2 29-May-2023 10:39:22
number of iterations : 3 29-May-2023 10:39:25
number of reformations : 3 29-May-2023 10:39:25
------- converged at time : 0.3 29-May-2023 10:39:25
number of iterations : 3 29-May-2023 10:39:28
number of reformations : 3 29-May-2023 10:39:28
------- converged at time : 0.4 29-May-2023 10:39:28
number of iterations : 3 29-May-2023 10:39:28
number of reformations : 3 29-May-2023 10:39:28
------- converged at time : 0.5 29-May-2023 10:39:28
number of iterations : 3 29-May-2023 10:39:32
number of reformations : 3 29-May-2023 10:39:32
------- converged at time : 0.6 29-May-2023 10:39:32
number of iterations : 3 29-May-2023 10:39:32
number of reformations : 3 29-May-2023 10:39:32
------- converged at time : 0.7 29-May-2023 10:39:32
number of iterations : 3 29-May-2023 10:39:34
number of reformations : 3 29-May-2023 10:39:34
------- converged at time : 0.8 29-May-2023 10:39:34
number of iterations : 3 29-May-2023 10:39:34
number of reformations : 3 29-May-2023 10:39:34
------- converged at time : 0.9 29-May-2023 10:39:34
number of iterations : 3 29-May-2023 10:39:36
number of reformations : 3 29-May-2023 10:39:36
------- converged at time : 1 29-May-2023 10:39:36
Elapsed time : 0:00:17 29-May-2023 10:39:36
N O R M A L T E R M I N A T I O N
# Done 29-May-2023 10:39:36
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
Import FEBio results
if runFlag==1 %i.e. a succesful run
Importing nodal displacements from a log file
dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_disp),0,1);
%Access data
N_disp_mat=dataStruct.data; %Displacement
timeVec=dataStruct.time; %Time
%Create deformed coordinate set
V_DEF=N_disp_mat+repmat(V,[1 1 size(N_disp_mat,3)]);
Importing element strain energies from a log file
dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_strainEnergy),0,1); %Element strain energy %Access data E_energy=dataStruct.data;
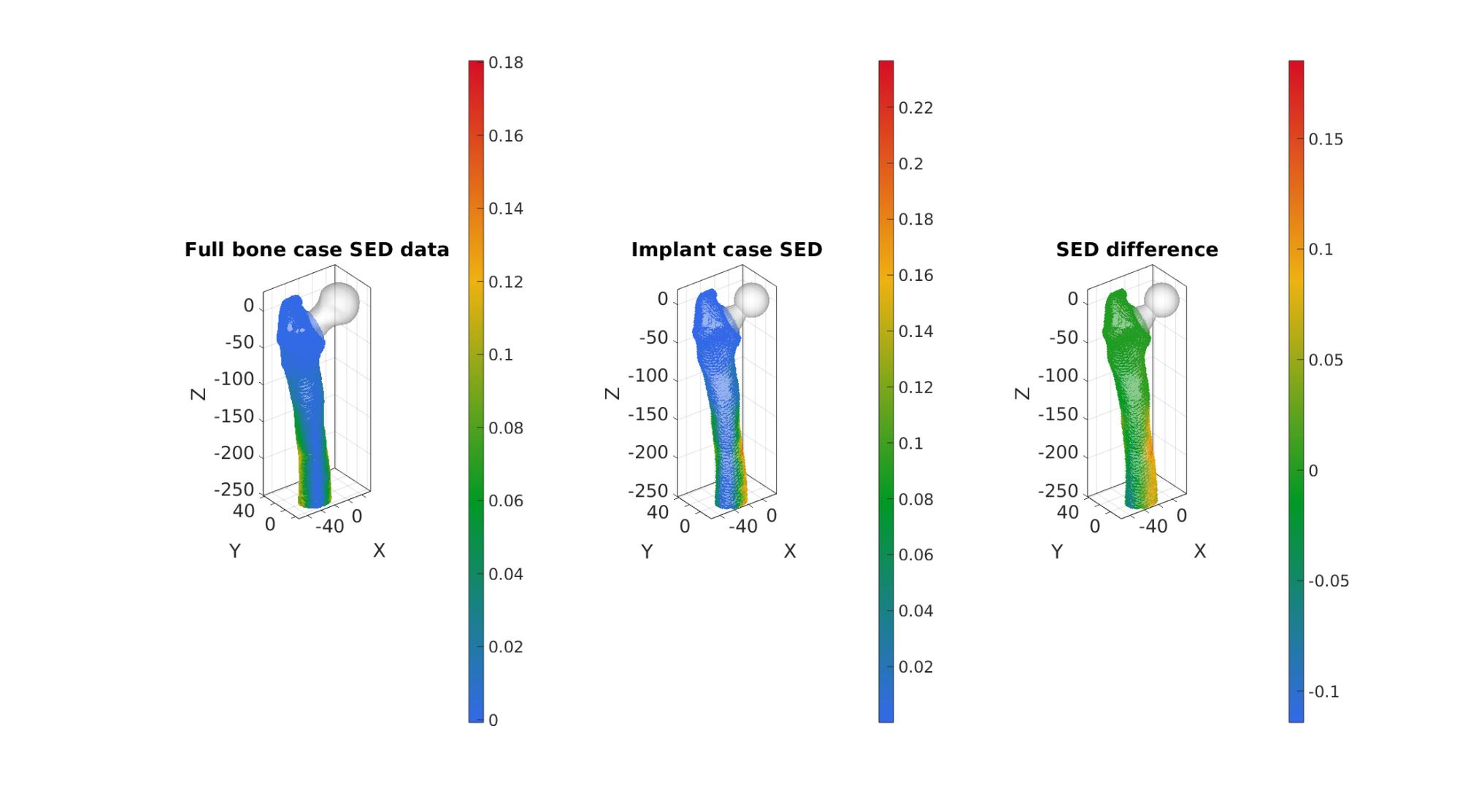
Compare to non-implant case
if skipCompare==0
% Interpolate at coordinates with implant W_p_at_VE_12=interpFuncEnergy(VE_12); W_12=E_energy(logicBoneElements,:,end); %SED for all elements delta_W=W_12-W_p_at_VE_12; %Energy difference between cases %Sum of squared delta_W delta_W_ssqd=sum(delta_W.^2);
cFigure;
subplot(1,3,1); hold on;
title('Full bone case SED data');
gpatch(Fb_no_implant,V_no_implant,'w','none',0.5); %Add graphics object to animate
scatterV(VE_12,50,W_p_at_VE_12,'filled');
axisGeom(gca,fontSize); colormap(gjet(250)); colorbar;
camlight headlight;
subplot(1,3,2); hold on;
title('Implant case SED');
gpatch(Fb,V,'w','none',0.5); %Add graphics object to animate
scatterV(VE_12,25,W_12,'filled');
axisGeom(gca,fontSize); colormap(gjet(250)); colorbar;
camlight headlight;
subplot(1,3,3); hold on;
title('SED difference');
gpatch(Fb,V,'w','none',0.5); %Add graphics object to animate
% gpatch(Fb_p,V_p,'w','none',0.5); %Add graphics object to animate
scatterV(VE_12,25,delta_W,'filled');
axisGeom(gca,fontSize); colormap(gjet(250)); colorbar;
camlight headlight;
drawnow;
C_data=delta_W;
n=[0 1 0]; %Normal direction to plane
P=mean(V,1); %Point on plane
[logicAt]=meshCleave(E_12,V,P,n);
% Get faces and matching color data for visualization
[F_cleave,CF_cleave]=element2patch(E_12(logicAt,:),C_data(logicAt));
hf=cFigure; hold on;
title('SED difference');
gpatch(Fb,V,'w','none',0.1); %Add graphics object to animate
hp1=gpatch(F_cleave,V,CF_cleave,'k',1);
axisGeom(gca,fontSize);
colormap(warmcold(250));
caxis([-(max(abs(C_data))) (max(abs(C_data)))]/2);
axis(axisLim(V_DEF)); %Set axis limits statically
colorbar; axis manual;
camlight headligth;
gdrawnow;
nSteps=25; %Number of animation steps
%Create the time vector
animStruct.Time=linspace(0,1,nSteps);
%The vector lengths
y=linspace(min(V(:,2)),max(V(:,2)),nSteps);
for q=1:1:nSteps
logicAt=meshCleave(E_12,V,[0 y(q) 0],n,[1 0]);
% Get faces and matching color data for cleaves elements
[F_cleave,CF_cleave]=element2patch(E_12(logicAt,:),C_data(logicAt));
%Set entries in animation structure
animStruct.Handles{q}=[hp1 hp1]; %Handles of objects to animate
animStruct.Props{q}={'Faces','CData'}; %Properties of objects to animate
animStruct.Set{q}={F_cleave,CF_cleave}; %Property values for to set in order to animate
end
anim8(hf,animStruct);
end
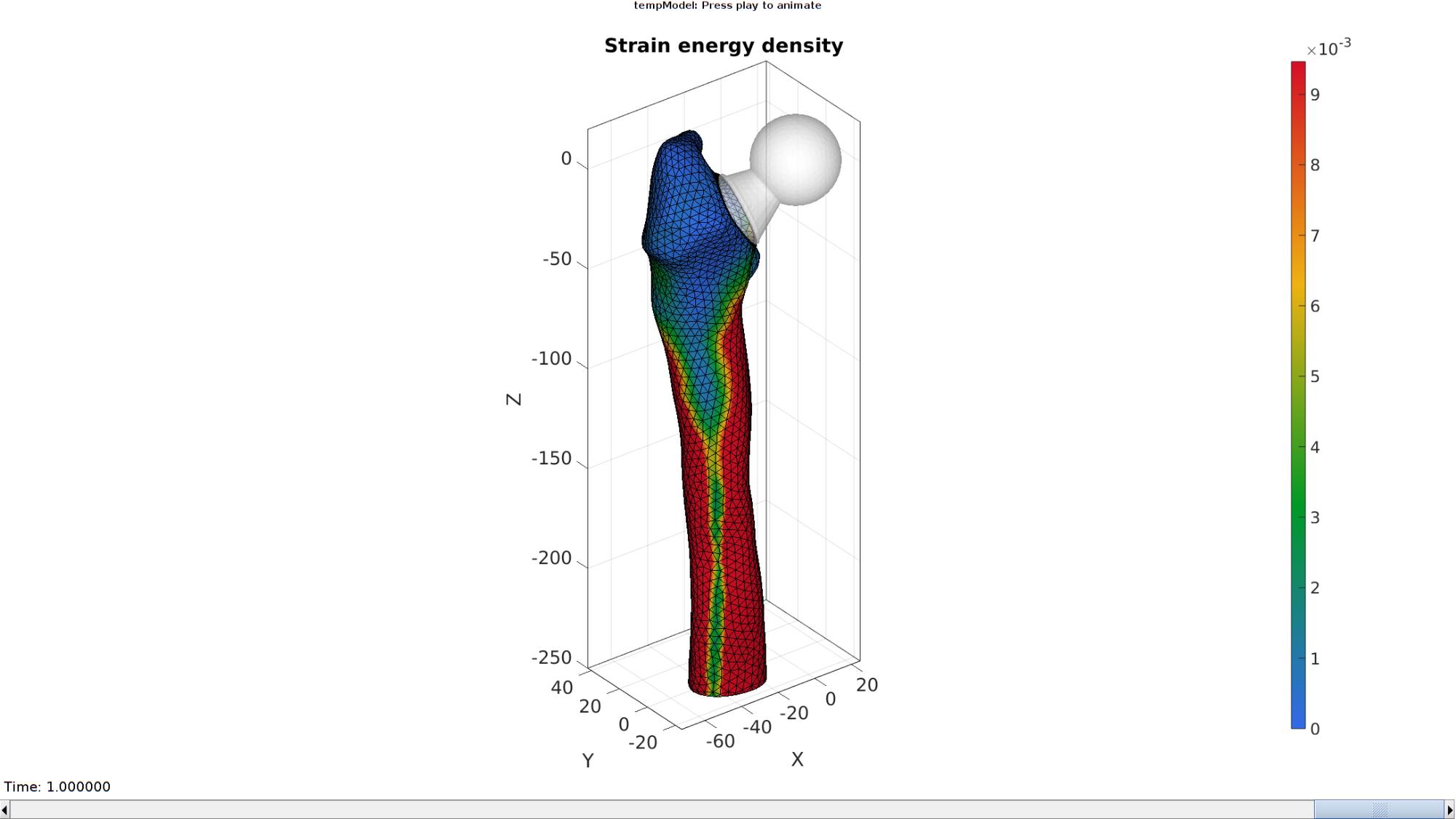
simulated results using anim8 to visualize and animate
deformations
FE_face=element2patch(E_12,[],'tet4'); [indBoundary]=tesBoundary(FE_face); Fb_energy=FE_face(indBoundary,:); [CV]=faceToVertexMeasure(E_12,V,E_energy(logicBoneElements,:,end)); % Create basic view and store graphics handle to initiate animation hf=cFigure; %Open figure title('Strain energy density') gtitle([febioFebFileNamePart,': Press play to animate']); hp1=gpatch(Fb(Cb>3,:),V_DEF(:,:,end),'w','none',0.5); %Add graphics object to animate hp2=gpatch(Fb_energy,V_DEF(:,:,end),CV,'k',1); %Add graphics object to animate hp2.FaceColor='Interp'; axisGeom(gca,fontSize); colormap(gjet(250)); colorbar; caxis([0 max(E_energy(:))/25]); axis(axisLim(V_DEF)); %Set axis limits statically camlight headlight; % Set up animation features animStruct.Time=timeVec; %The time vector for qt=1:1:size(N_disp_mat,3) %Loop over time increments DN=N_disp_mat(:,:,qt); %Current disp [CV]=faceToVertexMeasure(E_12,V,E_energy(logicBoneElements,:,qt)); %Set entries in animation structure animStruct.Handles{qt}=[hp1 hp2 hp2]; %Handles of objects to animate animStruct.Props{qt}={'Vertices','Vertices','CData'}; %Properties of objects to animate animStruct.Set{qt}={V_DEF(:,:,qt),V_DEF(:,:,qt),CV}; %Property values for to set in order to animate end anim8(hf,animStruct); %Initiate animation feature drawnow;
end
GIBBON footer text
License: https://github.com/gibbonCode/GIBBON/blob/master/LICENSE
GIBBON: The Geometry and Image-based Bioengineering add-On. A toolbox for image segmentation, image-based modeling, meshing, and finite element analysis.
Copyright (C) 2006-2023 Kevin Mattheus Moerman and the GIBBON contributors
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.