DEMO_febio_0060_vertebrae_disc_01
Below is a demonstration for:
- Building triangulated surface geometry for a spine segment
- Defining the boundary conditions
- Coding the febio structure
- Running the model
- Importing and visualizing results
Contents
- Keywords
- Control parameters
- Get vertebra bone model
- Remeshing
- Find top and bottom surface
- Get top and bottom boundary curves
- Resample meshes so curves end up with the same number of points
- Fix curve order
- Mesh disc with tetrahedral elements
- Joining node sets
- Define boundary conditions
- Defining the FEBio input structure
- Quick viewing of the FEBio input file structure
- Exporting the FEBio input file
- Running the FEBio analysis
- Import FEBio results
Keywords
- febio_spec version 4.0
- febio, FEBio
- spine, vertebra, disc
- contact, sliding, friction
- tetrahedral elements, tet4
- hexahedral elements, hex8
- static, solid
- hyperelastic, Ogden
- displacement logfile
clear; close all; clc;
Plot settings
fontSize=15; faceAlpha1=1; faceAlpha2=0.3; markerSize1=25; markerSize2=10; lineWidth=2; boneColor=[1.0000 0.9500 0.8000]; discColor=[0.8500 0.5000 0.3000];
Control parameters
% Path names defaultFolder = fileparts(fileparts(mfilename('fullpath'))); savePath=fullfile(defaultFolder,'data','temp'); % Defining file names febioFebFileNamePart='tempModel'; febioFebFileName=fullfile(savePath,[febioFebFileNamePart,'.feb']); %FEB file name febioLogFileName=fullfile(savePath,[febioFebFileNamePart,'.txt']); %FEBio log file name febioLogFileName_disp=[febioFebFileNamePart,'_disp_out.txt']; %Log file name for exporting displacement febioLogFileName_force=[febioFebFileNamePart,'_force_out.txt']; %Log file name for exporting force febioLogFileName_strainEnergy=[febioFebFileNamePart,'_energy_out.txt']; %Log file name for exporting strain energy density %Geometric parameters vertebraOffset=[0 0 30]; discHeight=12; rotAngleVert2=0; volumeFactorDisc=2; pointSpacing=3; %Approximate/desired node spacing annulusFibrosusFraction=0.5; numLayers=3; % Material parameters % Disc c1_1=1; %Shear-modulus-like parameter in MPa m1_1=2; %Material parameter setting degree of non-linearity k_1=c1_1*100; %Bulk modulus % FEA control settings numTimeSteps=10; %Number of time steps desired max_refs=25; %Max reforms max_ups=0; %Set to zero to use full-Newton iterations opt_iter=10; %Optimum number of iterations max_retries=8; %Maximum number of retires dtmin=(1/numTimeSteps)/100; %Minimum time step size dtmax=1/numTimeSteps; %Maximum time step size symmetric_stiffness=0; min_residual=1e-20; runMode='external'; %Boundary condition parameters forceApplied=1; displacementMagnitude=-1;
Get vertebra bone model
[F1,V1]=graphicsModels('vertebra');
rotAngle=-6;
R=euler2DCM([0 (rotAngle./180)*pi 0]);
V1=V1*R;
Remeshing
optionStructRemesh.pointSpacing=pointSpacing; %Set desired point spacing optionStructRemesh.disp_on=1; % Turn off command window text display [F1,V1]=ggremesh(F1,V1,optionStructRemesh);
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% ------> Geogram/vorpalite for resmeshing <------ 27-Apr-2023 16:51:09 # Export mesh input file. 27-Apr-2023 16:51:09 # Run Geomgram/vorpalite. 27-Apr-2023 16:51:09 ______________________________________________________________________________ | | | o-[config ] Configuration file name:geogram.ini | | Home directory:/home/kevin | | o-[I/O ] Output = /mnt/data/MATLAB/GIBBON/data/temp/temp_out.obj | | Loading file /mnt/data/MATLAB/GIBBON/data/temp/temp.obj... | | (FP64) nb_v:7500 nb_e:0 nb_f:15000 nb_b:0 tri:1 dim:3 | | Attributes on vertices: point[3] | | o-[Load ] Elapsed time: 0.01 s | ___________________________ _/ =====[preprocessing]===== \________________________________________________ | | | o-[CmdLine ] using pre:epsilon=0(0%) | | using pre:max_hole_area=100 | | using pre:min_comp_area=369.182(3%) | | o-[Components ] Nb connected components=1 | | Mesh does not have small connected component (good) | | o-[Validate ] Mesh does not have 0-area facets (good) | | o-[CmdLine ] using pre:margin=0(0%) | | o-[Pre ] Elapsed time: 0 s | _______________________ _/ =====[remeshing]===== \____________________________________________________ | | ||| o-[Newton ] Elapsed time: 0.18s | | o-[Remesh ] Computing RVD... | | o-[Validate ] (FP64) nb_v:1580 nb_e:0 nb_f:3160 nb_b:0 tri:1 dim:3 | | Attributes on vertices: point[3] | | o-[Remesh ] Elapsed time: 0.25 s | ____________________________ _/ =====[postprocessing]===== \_______________________________________________ | | | o-[CmdLine ] using post:min_comp_area=369.677(3%) | | o-[Components ] Nb connected components=1 | | Mesh does not have small connected component (good) | | o-[CmdLine ] using post:max_hole_area=100 | | using post:max_deg3_dist=0.122894(0.10000000000000001%) | | o-[Degree3 ] Does not have any degree 3 vertex (good) | | o-[Post ] Elapsed time: 0 s | ____________________ _/ =====[result]===== \_______________________________________________________ | | | o-[FinalMesh ] (FP64) nb_v:1580 nb_e:0 nb_f:3160 nb_b:0 tri:1 dim:3 | | Attributes on vertices: point[3] | | o-[I/O ] Saving file /mnt/data/MATLAB/GIBBON/data/temp/temp_out.obj. | | .. | | o-[Total time ] Elapsed time: 0.44 s | \______________________________________________________________________________/ # Importing remeshed geometry. 27-Apr-2023 16:51:09 # Removing temporary files. 27-Apr-2023 16:51:10 # Done! 27-Apr-2023 16:51:10
Find top and bottom surface
The below is a very basic hardcoded approach to finding (approximately) the top and bottom surfaces where the intevertebral discs would attached to the vertebra. The "detection" is based on the angle elements face with respect to the z-axis, and the distance from the origin (which lies in the middle in terms of the z-direction and in the centre of the disc attachement area in terms of the xy-direction).
angularThreshold=(25/180)*pi; distanceThreshold=26; V1F=patchCentre(F1,V1); N=patchNormal(F1,V1); nz=[0 0 1]; a=acos(dot(N,nz(ones(size(N,1),1),:),2)); D = sqrt(sum(V1F(:,[1 2]).^2,2)); logicTop = D<distanceThreshold & a<angularThreshold; indNotInLogic = unique(F1(~logicTop,:)); logicTop = logicTop & ~any(ismember(F1,indNotInLogic),2); logicTop = triSurfLogicSharpFix(F1,logicTop,3); D = sqrt(sum(V1F(:,[1 2]).^2,2)); logicBottom = D<distanceThreshold & a>(pi-angularThreshold); indNotInLogic = unique(F1(~logicBottom,:)); logicBottom = logicBottom & ~any(ismember(F1,indNotInLogic),2); logicBottom = triSurfLogicSharpFix(F1,logicBottom,3); C1=zeros(size(F1,1),1); C1(logicTop)=1; C1(logicBottom)=2;
cFigure; hold on; gpatch(F1,V1,C1,'k',1); axisGeom(gca,fontSize); colormap gjet; icolorbar; camlight('headlight'); gdrawnow;

Get top and bottom boundary curves
Eb1=patchBoundary(F1(C1==1,:)); indList1=edgeListToCurve(Eb1); indList1=indList1(1:end-1); Eb2=patchBoundary(F1(C1==2,:)); indList2=edgeListToCurve(Eb2); indList2=indList2(1:end-1);
Resample meshes so curves end up with the same number of points
n=[numel(indList1) numel(indList2)]; [nMax,indMax]=max(n); nn=abs(diff(n)); switch indMax case 1 [F1,V1,Eb2,C1]=triSurfSplitBoundary(F1,V1,Eb2,size(Eb2,1)+nn,C1); indList2=edgeListToCurve(Eb2); indList2=indList2(1:end-1); case 2 [F1,V1,Eb1,C1]=triSurfSplitBoundary(F1,V1,Eb1,size(Eb1,1)+nn,C1); indList1=edgeListToCurve(Eb1); indList1=indList1(1:end-1); end
Fix curve order
[~,indMin]=minDist(V1(indList1(1),:),V1(indList2,:)); if indMin>1 indList2=[indList2(indMin:end) indList2(1:indMin-1)]; end
nc=10; ns=50; [V1]=smoothCurve(F1,V1,nc,ns,indList1); [V1]=smoothCurve(F1,V1,nc,ns,indList2); P11=mean(V1(indList1,:),1); N11=mean(patchNormal(F1(C1==1,:),V1),1); P12=mean(V1(indList2,:),1); N12=-mean(patchNormal(F1(C1==2,:),V1),1);
cFigure; hold on; gpatch(F1,V1,C1,'k',1); % patchNormPlot(F1,V1) plotV(V1(indList1,:),'r.-','MarkerSize',25,'LineWidth',3); plotV(V1(indList2,:),'r.-','MarkerSize',25,'LineWidth',3); plotV(V1(indList1(1:2),:),'y.','MarkerSize',35); plotV(V1(indList2(1:2),:),'y.','MarkerSize',35); % plotV(V1(indRigid,:),'c.','MarkerSize',15); quiverVec(P11,N11,5,'r'); quiverVec(P12,N12,5,'r'); axisGeom(gca,fontSize); colormap gjet; icolorbar; camlight('headlight'); gdrawnow;
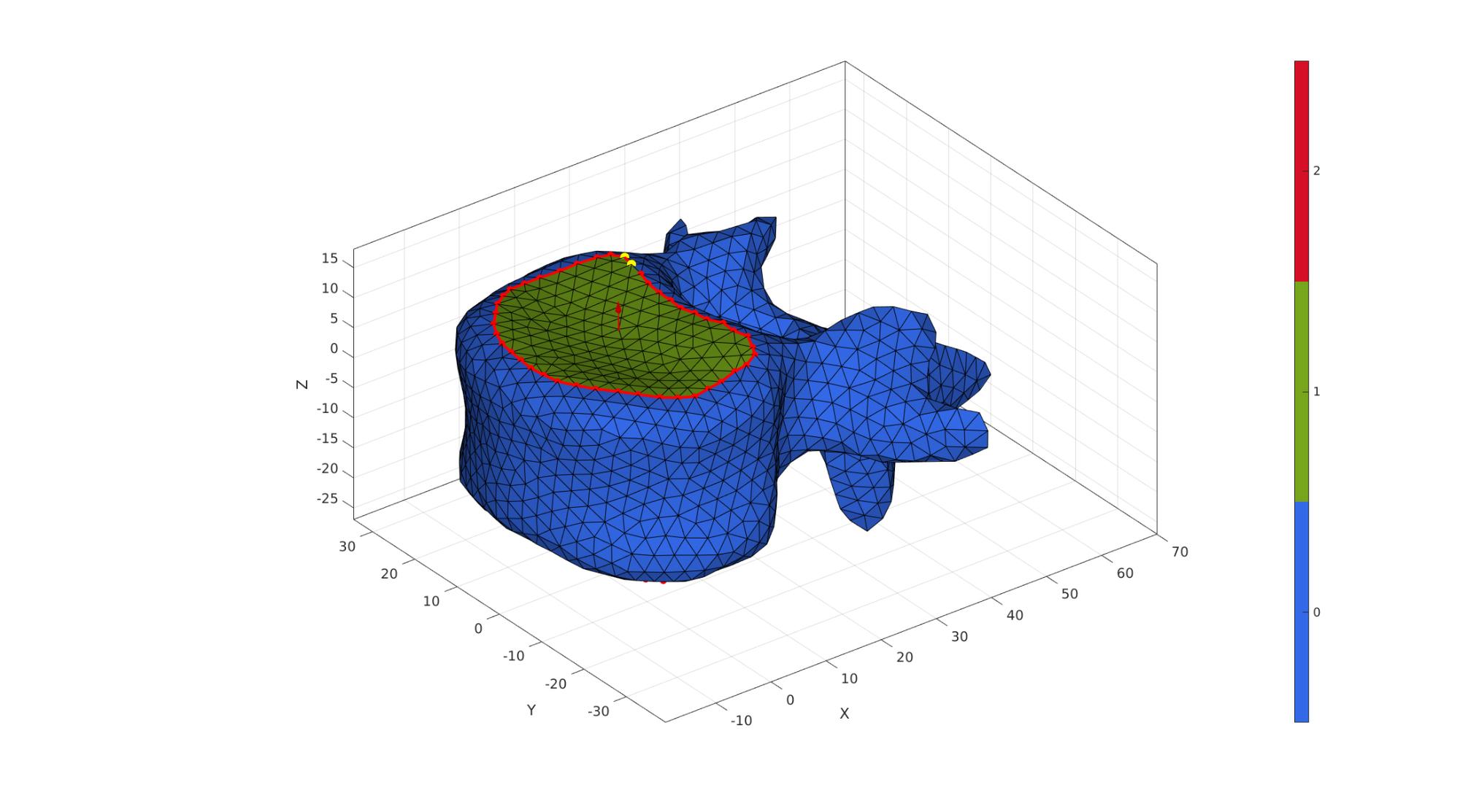
F2=F1; V2=V1; C2=C1; R2=euler2DCM([0 (rotAngleVert2./180)*pi 0]); V2=V2*R2; V2=V2+vertebraOffset(ones(size(V1,1),1),:); V2(:,3)=V2(:,3)+discHeight; P21=mean(V2(indList1,:),1); N21=mean(patchNormal(F2(C2==1,:),V2),1); P22=mean(V2(indList2,:),1); N22=-mean(patchNormal(F2(C2==2,:),V2),1);
f=discHeight/3; p=[P11;P11+f*N11; P22-f*N22; P22]; numStepsCurve=ceil(discHeight./pointSpacing); Vg=bezierCurve(p,numStepsCurve); [Fds,Vds,Cds]=sweepLoft(V1(indList1,:),V2(indList2,:),N11,N12,Vg,numStepsCurve); [~,~,Nd]=patchNormal(Fds,Vds); CVds=faceToVertexMeasure(Fds,Vds,Cds); CVds=CVds-min(CVds(:)); CVds=CVds./max(CVds(:)); CVds=abs(CVds-0.5); CVds=CVds./max(CVds(:)); CVds=CVds.^2; CVds=1-CVds; Vds=Vds+discHeight/10*Nd.*CVds; [Fds,Vds]=quad2tri(Fds,Vds,'a'); clear cPar; cPar.n=5; cPar.Method='HC'; cPar.RigidConstraints=unique(patchBoundary(Fds)); [Vds]=patchSmooth(Fds,Vds,[],cPar); [Fd,Vd,Cd]=joinElementSets({Fds,fliplr(F2(C2==2,:)),fliplr(F1(C1==1,:))},{Vds,V2,V1}); [Fd,Vd]=patchCleanUnused(Fd,Vd); [Fd,Vd]=mergeVertices(Fd,Vd);
Visualize imported surfaces
cFigure; hold on; gpatch(F1,V1,'rw','none',0.5); gpatch(F2,V2,'gw','none',0.5); gpatch(Fd,Vd,Cd,'k',1); % plotV(Vg,'r.-','MarkerSize',25,'LineWidth',3); camlight('headlight'); axisGeom(gca,fontSize); colormap gjet; icolorbar; gdrawnow;

Mesh disc with tetrahedral elements
Tet meshing is based on tetgen. TetGen requires a interior points for regions to be meshed, as well as intertior points for holes.
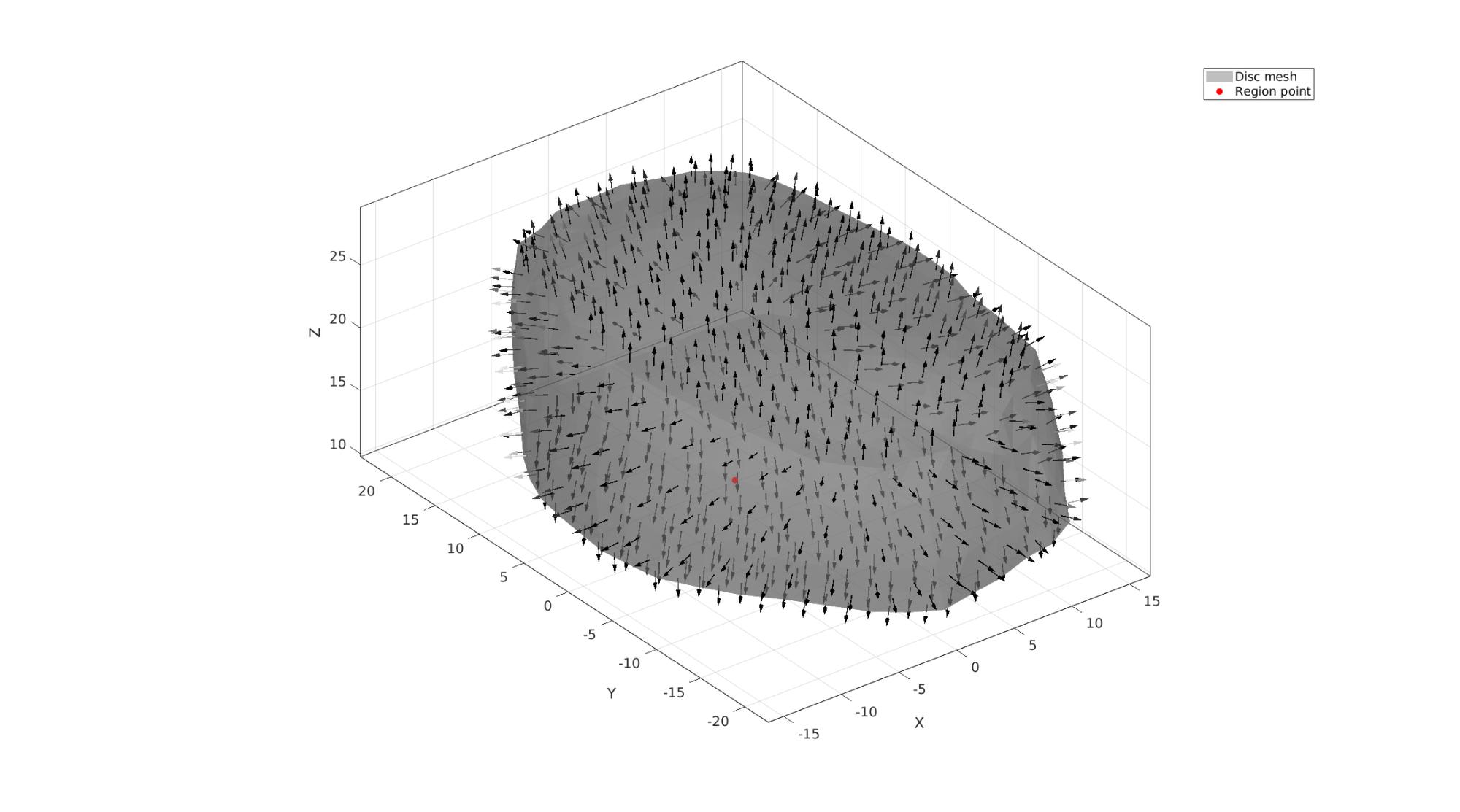
Define region points
[V_region]=getInnerPoint(Fd,Vd);
Visualize interior points
cFigure; hold on; hp1=gpatch(Fd,Vd,'kw','none',0.5); patchNormPlot(Fd,Vd); hp2=plotV(V_region,'r.','markerSize',markerSize1); legend([hp1 hp2],{'Disc mesh','Region point'}); axisGeom(gca,fontSize); camlight('headlight'); gdrawnow;
Mesh using tetgen
inputStruct.stringOpt='-pq1.2AaY'; %TetGen option string inputStruct.Faces=Fd; %The faces inputStruct.Nodes=Vd; %The vertices inputStruct.holePoints=[]; %The hole interior points inputStruct.faceBoundaryMarker=Cd; %Face boundary markers inputStruct.regionPoints=V_region; %The region interior points inputStruct.regionA=tetVolMeanEst(Fd,Vd)*volumeFactorDisc; %Volume for regular tets
Mesh model using tetrahedral elements using tetGen
[meshOutput]=runTetGen(inputStruct); %Run tetGen
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- TETGEN Tetrahedral meshing --- 27-Apr-2023 16:51:16 %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- Writing SMESH file --- 27-Apr-2023 16:51:16 ----> Adding node field ----> Adding facet field ----> Adding holes specification ----> Adding region specification --- Done --- 27-Apr-2023 16:51:17 --- Running TetGen to mesh input boundary--- 27-Apr-2023 16:51:17 Opening /mnt/data/MATLAB/GIBBON/data/temp/temp.smesh. Delaunizing vertices... Delaunay seconds: 0.001373 Creating surface mesh ... Surface mesh seconds: 0.000537 Recovering boundaries... Boundary recovery seconds: 0.000744 Removing exterior tetrahedra ... Spreading region attributes. Exterior tets removal seconds: 0.000479 Recovering Delaunayness... Delaunay recovery seconds: 0.00041 Refining mesh... 515 insertions, added 310 points, 246 tetrahedra in queue. Refinement seconds: 0.008608 Smoothing vertices... Mesh smoothing seconds: 0.012225 Improving mesh... Mesh improvement seconds: 0.000696 Writing /mnt/data/MATLAB/GIBBON/data/temp/temp.1.node. Writing /mnt/data/MATLAB/GIBBON/data/temp/temp.1.ele. Writing /mnt/data/MATLAB/GIBBON/data/temp/temp.1.face. Writing /mnt/data/MATLAB/GIBBON/data/temp/temp.1.edge. Output seconds: 0.007349 Total running seconds: 0.032498 Statistics: Input points: 387 Input facets: 770 Input segments: 1155 Input holes: 0 Input regions: 1 Mesh points: 707 Mesh tetrahedra: 3056 Mesh faces: 6497 Mesh faces on exterior boundary: 770 Mesh faces on input facets: 770 Mesh edges on input segments: 1155 Steiner points inside domain: 320 --- Done --- 27-Apr-2023 16:51:17 %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- Importing TetGen files --- 27-Apr-2023 16:51:17 --- Done --- 27-Apr-2023 16:51:17
Access model element and patch data
Fb_disc=meshOutput.facesBoundary; %Boundary faces of the disc Cb_disc=meshOutput.boundaryMarker; %Boundary marker/color data for the disc V_disc=meshOutput.nodes; %The vertices/nodes E_disc=meshOutput.elements; %The tet4 elements
Visualizing mesh using meshView, see also anim8
meshView(meshOutput);

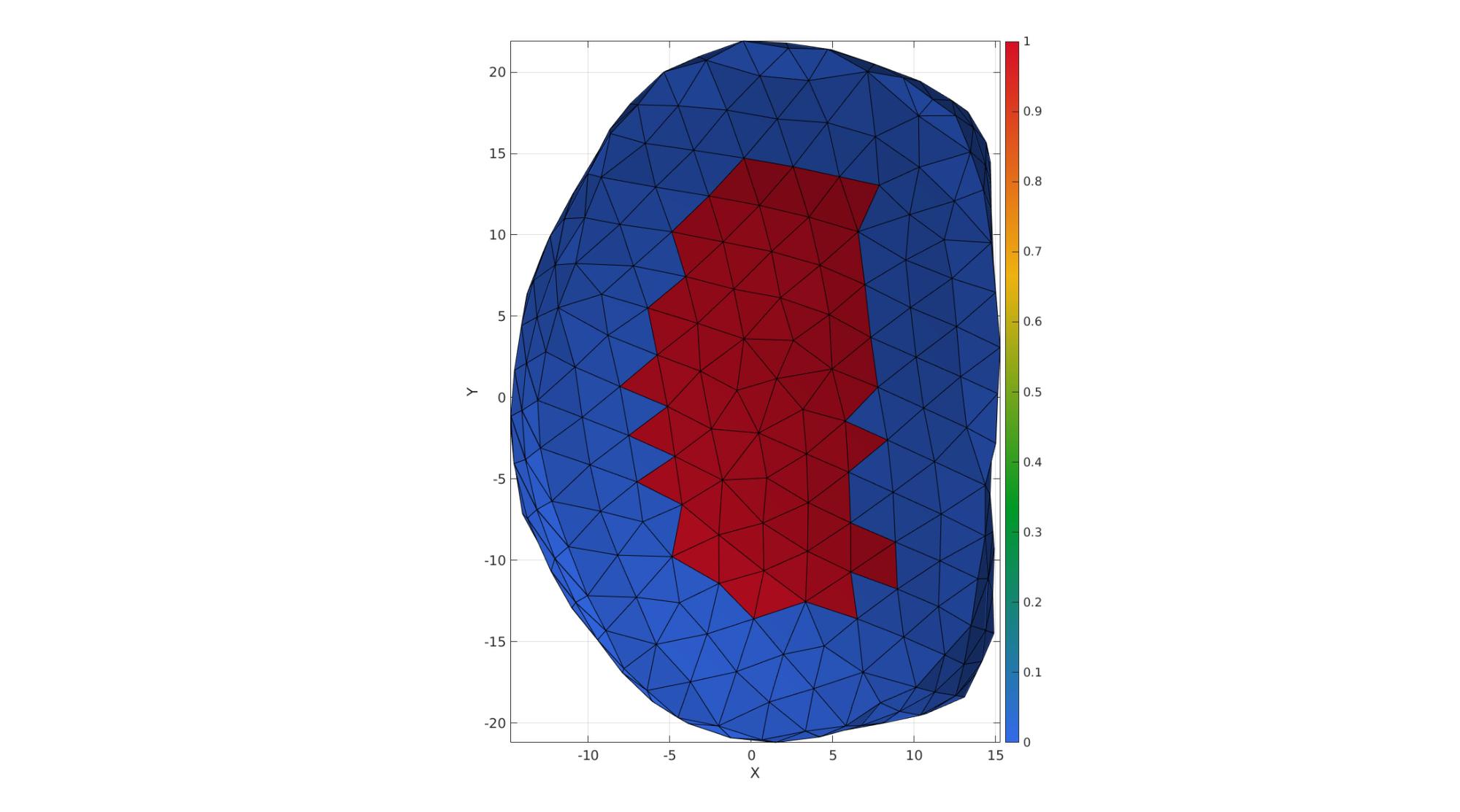
VE_disc=patchCentre(E_disc,V_disc); Eb=patchBoundary(Fb_disc(ismember(Cb_disc,[2,3]),:)); indB=unique(Fb_disc(ismember(Cb_disc,1),:)); [D,~]=minDist(VE_disc(:,[1 2]),V_disc(indB,[1 2])); D=D./max(D(:)); D=1-D; logicAnnulusFibrosus=D<=annulusFibrosusFraction; D(logicAnnulusFibrosus)=annulusFibrosusFraction; D=D-min(D(:)); D=D./max(D(:)); [FE,CF]=element2patch(E_disc,logicAnnulusFibrosus);
cFigure; hold on; gpatch(FE,V_disc,CF,'k',1); axisGeom; camlight headlight; view(2); colormap gjet; colorbar; gdrawnow;
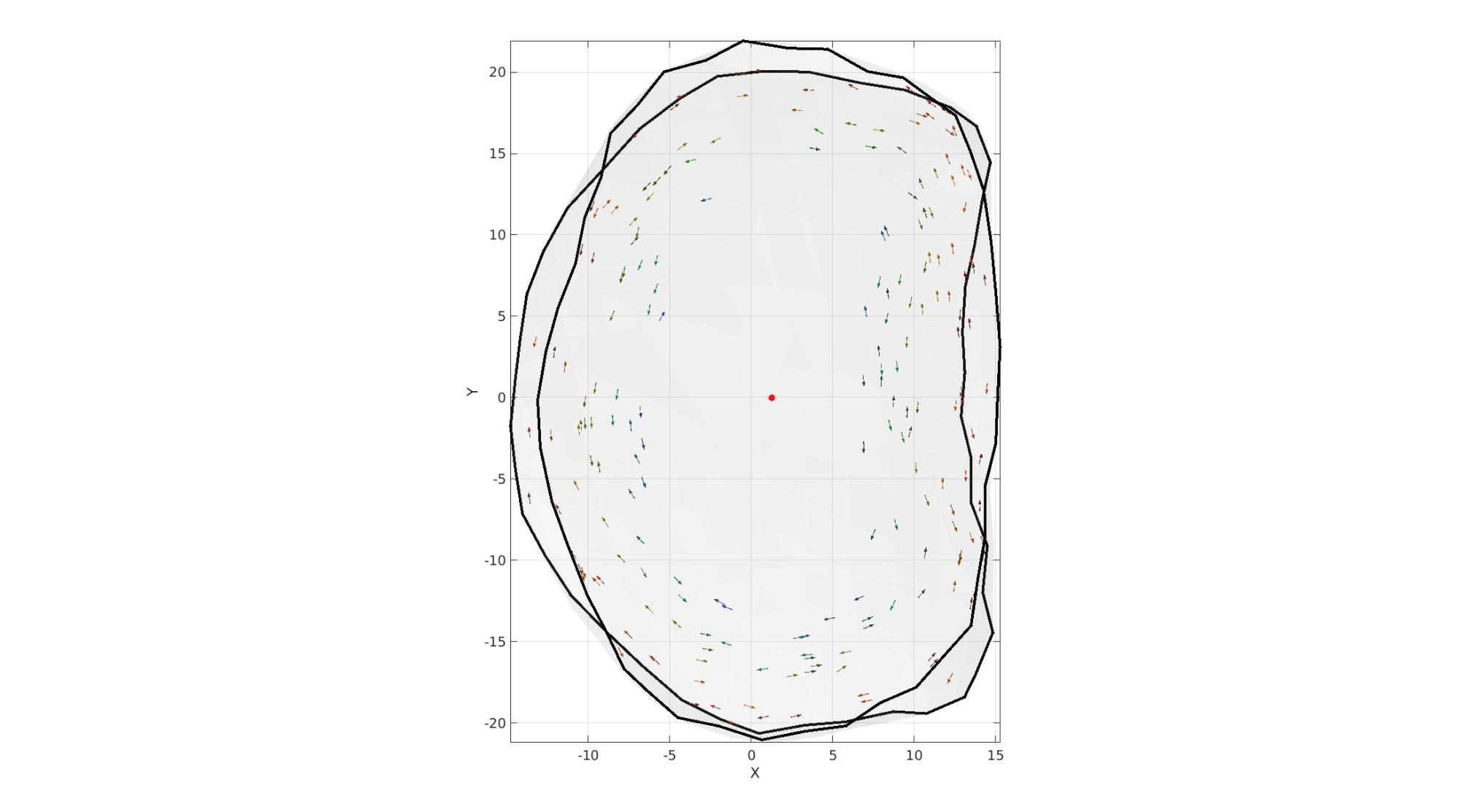
VEb=patchCentre(Eb,V_disc); t = atan2(VEb(:,2),VEb(:,1)); T = atan2(VE_disc(:,2),VE_disc(:,1)); VEb=patchCentre(Eb,V_disc); NE=vecnormalize(V_disc(Eb(:,1),:)-V_disc(Eb(:,2),:)); NE(:,3)=0; NE=vecnormalize(NE); [~,indMin]=minDist(T,t); % z=[0 0 1]; % NFC=cross(NF,z(ones(size(NF,1),1),:),2); NE_disc=NE(indMin,:); % NE_disc(:,[1 2])=NE_disc(:,[1 2]).*sin(numLayers.*D(:,ones(1,2)).*2*pi); % NE_disc(:,3)=cos(numLayers.*D.*2*pi); % NE_disc(logicAnnulusFibrosus,:)=NaN; logicEven=iseven(round(D.*numLayers)); NE_disc(logicEven,3)=1; NE_disc(~logicEven,3)=-1; NE_disc=vecnormalize(NE_disc); NE_disc(logicAnnulusFibrosus,:)=NaN; cFigure; hold on; gpatch(Fb_disc,V_disc,'w','none',0.1); gpatch(Eb,V_disc,'none','k',1,3); % plotV(VE_disc,'k.','MarkerSize',1); plotV(mean(V_disc,1),'r.','MarkerSize',25); quiverVec(VE_disc(1:10:end,:),NE_disc(1:10:end,:),1,D(1:10:end,:)); axisGeom; camlight headlight; view(2); colormap gjet; gdrawnow;
Joining node sets
V=[V_disc;V1;V2]; %Combined node sets E1=F1+size(V_disc,1); %Fixed element indices E2=F2+size(V_disc,1)+size(V1,1); %Fixed element indices numDigitsMerge=6-numOrder(pointSpacing); [~,indKeep,indFix]=unique(pround(V,numDigitsMerge),'rows'); V=V(indKeep,:); E_disc=indFix(E_disc); E1=indFix(E1); E2=indFix(E2); Fb_disc=indFix(Fb_disc);
Define boundary conditions
Visualize BC's
cFigure; hold on; title('Boundary conditions'); hp(1)=gpatch(Fb_disc,V,'w','none',0.5); hp(2)=gpatch(E1,V,'rw','none',1); hp(3)=gpatch(E2,V,'bw','none',1); legend(hp,{'Disc','Constrained vertebra','BC vertebra'}); axisGeom; camlight headlight; gdrawnow;

Defining the FEBio input structure
See also febioStructTemplate and febioStruct2xml and the FEBio user manual.
%Get a template with default settings [febio_spec]=febioStructTemplate; %febio_spec version febio_spec.ATTR.version='4.0'; %Module section febio_spec.Module.ATTR.type='solid'; %Control section febio_spec.Control.analysis='STATIC'; febio_spec.Control.time_steps=numTimeSteps; febio_spec.Control.step_size=1/numTimeSteps; febio_spec.Control.solver.max_refs=max_refs; febio_spec.Control.solver.qn_method.max_ups=max_ups; febio_spec.Control.time_stepper.dtmin=dtmin; febio_spec.Control.time_stepper.dtmax=dtmax; febio_spec.Control.time_stepper.max_retries=max_retries; febio_spec.Control.time_stepper.opt_iter=opt_iter; %Material section materialName1='Material1'; febio_spec.Material.material{1}.ATTR.name=materialName1; febio_spec.Material.material{1}.ATTR.type='Ogden'; febio_spec.Material.material{1}.ATTR.id=1; febio_spec.Material.material{1}.c1=c1_1; febio_spec.Material.material{1}.m1=m1_1; febio_spec.Material.material{1}.c2=c1_1; febio_spec.Material.material{1}.m2=-m1_1; febio_spec.Material.material{1}.k=k_1; materialName2='Material2'; febio_spec.Material.material{2}.ATTR.name=materialName2; febio_spec.Material.material{2}.ATTR.type='rigid body'; febio_spec.Material.material{2}.ATTR.id=2; febio_spec.Material.material{2}.density=1; febio_spec.Material.material{2}.center_of_mass=mean(V(unique(E1),:),1); materialName3='Material3'; febio_spec.Material.material{3}.ATTR.name=materialName3; febio_spec.Material.material{3}.ATTR.type='rigid body'; febio_spec.Material.material{3}.ATTR.id=3; febio_spec.Material.material{3}.density=1; febio_spec.Material.material{3}.center_of_mass=mean(V(unique(E2),:),1); % Mesh section % -> Nodes febio_spec.Mesh.Nodes{1}.ATTR.name='Object1'; %The node set name febio_spec.Mesh.Nodes{1}.node.ATTR.id=(1:size(V,1))'; %The node id's febio_spec.Mesh.Nodes{1}.node.VAL=V; %The nodel coordinates % -> Elements partName1='Part1'; febio_spec.Mesh.Elements{1}.ATTR.name=partName1; %Name of this part febio_spec.Mesh.Elements{1}.ATTR.type='tet4'; %Element type febio_spec.Mesh.Elements{1}.elem.ATTR.id=(1:1:size(E_disc,1))'; %Element id's febio_spec.Mesh.Elements{1}.elem.VAL=E_disc; %The element matrix partName2='Part2'; febio_spec.Mesh.Elements{2}.ATTR.name=partName2; %Name of this part febio_spec.Mesh.Elements{2}.ATTR.type='tri3'; %Element type febio_spec.Mesh.Elements{2}.elem.ATTR.id=size(E_disc,1)+(1:1:size(E1,1))'; %Element id's febio_spec.Mesh.Elements{2}.elem.VAL=E1; %The element matrix partName3='Part3'; febio_spec.Mesh.Elements{3}.ATTR.name=partName3; %Name of this part febio_spec.Mesh.Elements{3}.ATTR.type='tri3'; %Element type febio_spec.Mesh.Elements{3}.elem.ATTR.id=size(E_disc,1)+size(E1,1)+(1:1:size(E2,1))'; %Element id's febio_spec.Mesh.Elements{3}.elem.VAL=E2; %The element matrix %MeshDomains section febio_spec.MeshDomains.SolidDomain{1}.ATTR.name=partName1; febio_spec.MeshDomains.SolidDomain{1}.ATTR.mat=materialName1; febio_spec.MeshDomains.ShellDomain{1}.ATTR.name=partName2; febio_spec.MeshDomains.ShellDomain{1}.ATTR.mat=materialName2; febio_spec.MeshDomains.ShellDomain{2}.ATTR.name=partName3; febio_spec.MeshDomains.ShellDomain{2}.ATTR.mat=materialName3; %Rigid section % ->Rigid body fix boundary conditions febio_spec.Rigid.rigid_bc{1}.ATTR.name='RigidFix1'; febio_spec.Rigid.rigid_bc{1}.ATTR.type='rigid_fixed'; febio_spec.Rigid.rigid_bc{1}.rb=2; febio_spec.Rigid.rigid_bc{1}.Rx_dof=1; febio_spec.Rigid.rigid_bc{1}.Ry_dof=1; febio_spec.Rigid.rigid_bc{1}.Rz_dof=1; febio_spec.Rigid.rigid_bc{1}.Ru_dof=1; febio_spec.Rigid.rigid_bc{1}.Rv_dof=1; febio_spec.Rigid.rigid_bc{1}.Rw_dof=1; febio_spec.Rigid.rigid_bc{2}.ATTR.name='RigidFix2'; febio_spec.Rigid.rigid_bc{2}.ATTR.type='rigid_fixed'; febio_spec.Rigid.rigid_bc{2}.rb=3; febio_spec.Rigid.rigid_bc{2}.Rx_dof=1; febio_spec.Rigid.rigid_bc{2}.Ry_dof=1; febio_spec.Rigid.rigid_bc{2}.Ru_dof=1; febio_spec.Rigid.rigid_bc{2}.Rv_dof=1; febio_spec.Rigid.rigid_bc{2}.Rw_dof=1; % ->Rigid body prescribe boundary conditions febio_spec.Rigid.rigid_bc{3}.ATTR.name='RigidPrescribe'; febio_spec.Rigid.rigid_bc{3}.ATTR.type='rigid_displacement'; febio_spec.Rigid.rigid_bc{3}.rb=3; febio_spec.Rigid.rigid_bc{3}.dof='z'; febio_spec.Rigid.rigid_bc{3}.value.ATTR.lc=1; febio_spec.Rigid.rigid_bc{3}.value.VAL=displacementMagnitude; febio_spec.Rigid.rigid_bc{3}.relative=0; %LoadData section % -> load_controller febio_spec.LoadData.load_controller{1}.ATTR.name='LC_1'; febio_spec.LoadData.load_controller{1}.ATTR.id=1; febio_spec.LoadData.load_controller{1}.ATTR.type='loadcurve'; febio_spec.LoadData.load_controller{1}.interpolate='LINEAR'; %febio_spec.LoadData.load_controller{1}.extend='CONSTANT'; febio_spec.LoadData.load_controller{1}.points.pt.VAL=[0 0; 1 1]; %Output section % -> log file febio_spec.Output.logfile.ATTR.file=febioLogFileName; febio_spec.Output.logfile.node_data{1}.ATTR.file=febioLogFileName_disp; febio_spec.Output.logfile.node_data{1}.ATTR.data='ux;uy;uz'; febio_spec.Output.logfile.node_data{1}.ATTR.delim=','; febio_spec.Output.logfile.node_data{2}.ATTR.file=febioLogFileName_force; febio_spec.Output.logfile.node_data{2}.ATTR.data='Rx;Ry;Rz'; febio_spec.Output.logfile.node_data{2}.ATTR.delim=','; febio_spec.Output.logfile.element_data{1}.ATTR.file=febioLogFileName_strainEnergy; febio_spec.Output.logfile.element_data{1}.ATTR.data='sed'; febio_spec.Output.logfile.element_data{1}.ATTR.delim=','; febio_spec.Output.logfile.element_data{1}.VAL=1:1:size(E_disc,1); % Plotfile section febio_spec.Output.plotfile.compression=0;
Quick viewing of the FEBio input file structure
The febView function can be used to view the xml structure in a MATLAB figure window.
febView(febio_spec); %Viewing the febio file
Exporting the FEBio input file
Exporting the febio_spec structure to an FEBio input file is done using the febioStruct2xml function.
febioStruct2xml(febio_spec,febioFebFileName); %Exporting to file and domNode
Running the FEBio analysis
To run the analysis defined by the created FEBio input file the runMonitorFEBio function is used. The input for this function is a structure defining job settings e.g. the FEBio input file name. The optional output runFlag informs the user if the analysis was run succesfully.
febioAnalysis.run_filename=febioFebFileName; %The input file name febioAnalysis.run_logname=febioLogFileName; %The name for the log file febioAnalysis.disp_on=1; %Display information on the command window febioAnalysis.runMode=runMode;%'internal'; [runFlag]=runMonitorFEBio(febioAnalysis);%START FEBio NOW!!!!!!!!
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
--------> RUNNING/MONITORING FEBIO JOB <-------- 27-Apr-2023 16:51:26
FEBio path: /home/kevin/FEBioStudio2/bin/febio4
# Attempt removal of existing log files 27-Apr-2023 16:51:26
* Removal succesful 27-Apr-2023 16:51:26
# Attempt removal of existing .xplt files 27-Apr-2023 16:51:26
* Removal succesful 27-Apr-2023 16:51:26
# Starting FEBio... 27-Apr-2023 16:51:26
Max. total analysis time is: Inf s
* Waiting for log file creation 27-Apr-2023 16:51:26
Max. wait time: 30 s
* Log file found. 27-Apr-2023 16:51:26
# Parsing log file... 27-Apr-2023 16:51:26
number of iterations : 3 27-Apr-2023 16:51:27
number of reformations : 3 27-Apr-2023 16:51:27
------- converged at time : 0.1 27-Apr-2023 16:51:27
number of iterations : 3 27-Apr-2023 16:51:27
number of reformations : 3 27-Apr-2023 16:51:27
------- converged at time : 0.2 27-Apr-2023 16:51:27
number of iterations : 3 27-Apr-2023 16:51:27
number of reformations : 3 27-Apr-2023 16:51:27
------- converged at time : 0.3 27-Apr-2023 16:51:27
number of iterations : 3 27-Apr-2023 16:51:28
number of reformations : 3 27-Apr-2023 16:51:28
------- converged at time : 0.4 27-Apr-2023 16:51:28
number of iterations : 3 27-Apr-2023 16:51:28
number of reformations : 3 27-Apr-2023 16:51:28
------- converged at time : 0.5 27-Apr-2023 16:51:28
number of iterations : 3 27-Apr-2023 16:51:28
number of reformations : 3 27-Apr-2023 16:51:28
------- converged at time : 0.6 27-Apr-2023 16:51:28
number of iterations : 3 27-Apr-2023 16:51:28
number of reformations : 3 27-Apr-2023 16:51:28
------- converged at time : 0.7 27-Apr-2023 16:51:28
number of iterations : 3 27-Apr-2023 16:51:28
number of reformations : 3 27-Apr-2023 16:51:28
------- converged at time : 0.8 27-Apr-2023 16:51:28
number of iterations : 3 27-Apr-2023 16:51:28
number of reformations : 3 27-Apr-2023 16:51:28
------- converged at time : 0.9 27-Apr-2023 16:51:28
number of iterations : 3 27-Apr-2023 16:51:28
number of reformations : 3 27-Apr-2023 16:51:28
------- converged at time : 1 27-Apr-2023 16:51:28
Elapsed time : 0:00:02 27-Apr-2023 16:51:28
N O R M A L T E R M I N A T I O N
# Done 27-Apr-2023 16:51:28
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
Import FEBio results
if runFlag==1 %i.e. a succesful run
Importing nodal displacements from a log file
dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_disp),0,1);
%Access data
N_disp_mat=dataStruct.data; %Displacement
timeVec=dataStruct.time; %Time
%Create deformed coordinate set
V_DEF=N_disp_mat+repmat(V,[1 1 size(N_disp_mat,3)]);
Importing element stress from a log file
dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_strainEnergy),0,1);
%Access data
E_sed_mat=dataStruct.data;
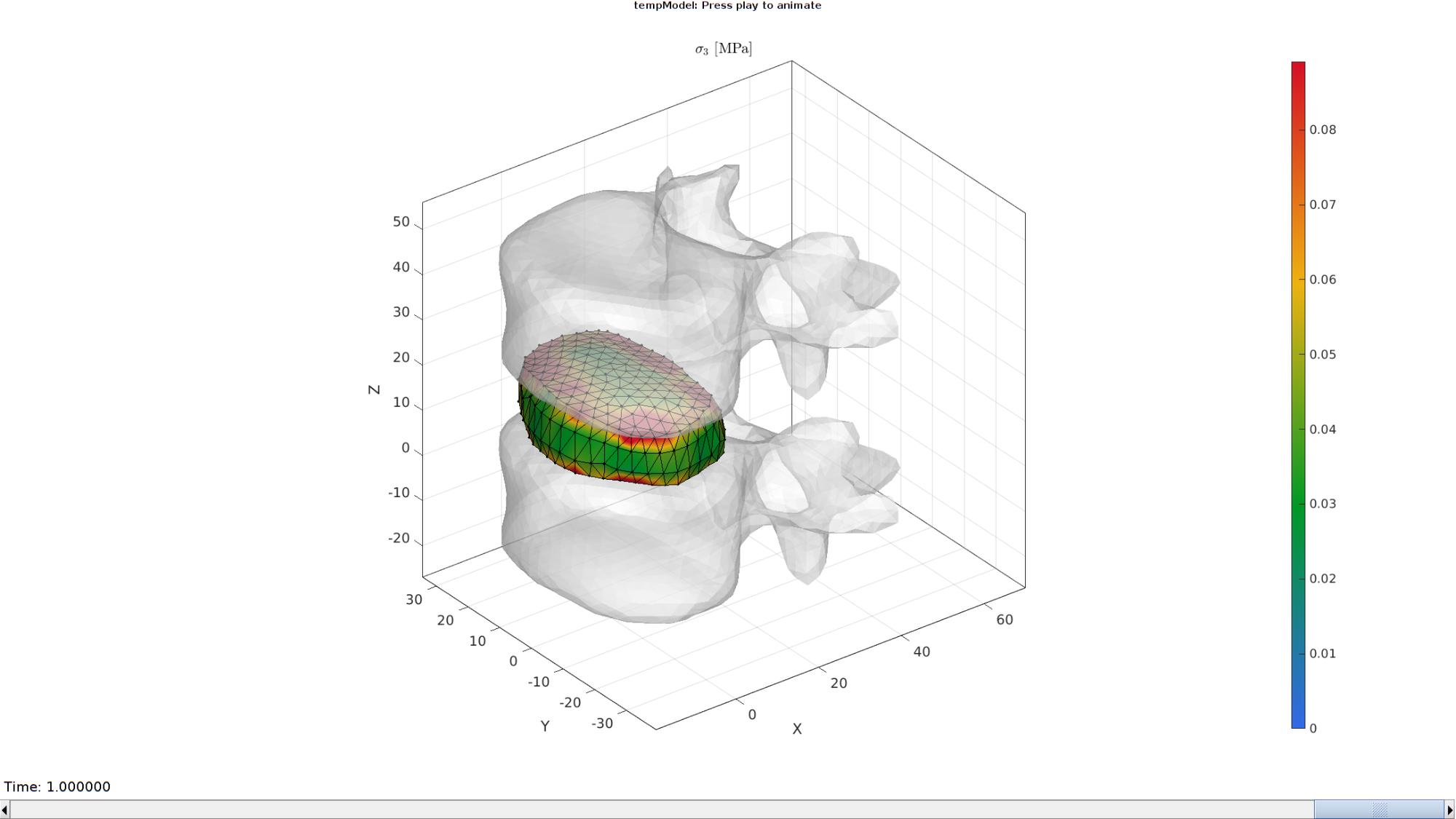
Plotting the simulated results using anim8 to visualize and animate deformations
[CV]=faceToVertexMeasure(E_disc,V,E_sed_mat(:,:,end));
% Create basic view and store graphics handle to initiate animation
hf=cFigure; %Open figure
gtitle([febioFebFileNamePart,': Press play to animate']);
title('$\sigma_{3}$ [MPa]','Interpreter','Latex')
hp=gpatch(Fb_disc,V_DEF(:,:,end),CV,'k',1); %Add graphics object to animate
hp.Marker='.';
hp.MarkerSize=markerSize2;
hp.FaceColor='interp';
hp2=gpatch([E1;E2],V_DEF(:,:,end),'w','none',0.5); %Add graphics object to animate
axisGeom(gca,fontSize);
colormap(gjet(250)); colorbar;
caxis([0 max(E_sed_mat(:))/10]);
axis(axisLim(V_DEF)); %Set axis limits statically
camlight headlight;
% Set up animation features
animStruct.Time=timeVec; %The time vector
for qt=1:1:size(N_disp_mat,3) %Loop over time increments
[CV]=faceToVertexMeasure(E_disc,V,E_sed_mat(:,:,qt));
%Set entries in animation structure
animStruct.Handles{qt}=[hp hp hp2]; %Handles of objects to animate
animStruct.Props{qt}={'Vertices','CData','Vertices'}; %Properties of objects to animate
animStruct.Set{qt}={V_DEF(:,:,qt),CV,V_DEF(:,:,qt)}; %Property values for to set in order to animate
end
anim8(hf,animStruct); %Initiate animation feature
drawnow;
end
function [V1]=smoothCurve(F1,V1,nc,ns,indList1) clear cPar cPar.n=nc; cPar.Method='HC'; e=[(1:numel(indList1))' [(2:numel(indList1))';1]]; v=V1(indList1,:); [v]=patchSmooth(e,v,[],cPar); V1(indList1,:)=v; indTouch=unique(F1(any(ismember(F1,indList1),2),:)); indTouch=unique(F1(any(ismember(F1,indTouch),2),:)); logicRigid=true(size(V1,1),1); logicRigid(indTouch)=0; logicRigid(indList1)=1; indRigid=find(logicRigid); cPar.n=ns; cPar.Method='HC'; cPar.RigidConstraints=indRigid; [V1]=patchSmooth(F1,V1,[],cPar); end

GIBBON www.gibboncode.org
Kevin Mattheus Moerman, [email protected]
GIBBON footer text
License: https://github.com/gibbonCode/GIBBON/blob/master/LICENSE
GIBBON: The Geometry and Image-based Bioengineering add-On. A toolbox for image segmentation, image-based modeling, meshing, and finite element analysis.
Copyright (C) 2006-2022 Kevin Mattheus Moerman and the GIBBON contributors
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.