DEMO_febio_0054_lattice_hydrostatic_01
Below is a demonstration for:
- Building geometry for a cube with hexahedral elements
- Defining the boundary conditions
- Coding the febio structure
- Running the model
- Importing and visualizing the displacement and stress results
Contents
Keywords
- febio_spec version 4.0
- febio, FEBio
- uniaxial loading
- compression, tension, compressive, tensile
- displacement control, displacement boundary condition
- hexahedral elements, hex8
- cube, box, rectangular
- static, solid
- hyperelastic, Ogden
- displacement logfile
- stress logfile
clear; close all; clc;
Plot settings
Plot settings
fontSize=15; faceAlpha1=0.8; faceAlpha2=1; edgeColor=0.25*ones(1,3); edgeWidth=1.5; markerSize=25; cMap=gjet(4);
Control parameters
% Path names defaultFolder = fileparts(fileparts(mfilename('fullpath'))); savePath=fullfile(defaultFolder,'data','temp'); % Defining file names febioFebFileNamePart='tempModel'; febioFebFileName=fullfile(savePath,[febioFebFileNamePart,'.feb']); %FEB file name febioLogFileName=fullfile(savePath,[febioFebFileNamePart,'.txt']); %FEBio log file name febioLogFileName_disp=[febioFebFileNamePart,'_disp_out.txt']; %Log file name for exporting displacement febioLogFileName_force=[febioFebFileNamePart,'_force_out.txt']; %Log file name for exporting force febioLogFileName_stress=[febioFebFileNamePart,'_stress_out.txt']; %Log file name for exporting stress febioLogFileName_stiffness=[febioFebFileNamePart,'_stiffness_out.txt']; %Log file name for exporting stiffness %Specifying dimensions and number of elements sampleSize=10; latticeType=1; %Define applied displacement J_final=0.7; %Final Jacobian or volume ration lambdaFinal=J_final^(1/3); %Stretch values in all directions displacementMagnitude=((lambdaFinal*sampleSize)-sampleSize)/2; %The displacement magnitude %Material parameter set c1=1; %Shear-modulus-like parameter m1=2; k=50*c1; % FEA control settings numTimeSteps=25; %Number of time steps desired max_refs=50; %Max reforms max_ups=0; %Set to zero to use full-Newton iterations opt_iter=6; %Optimum number of iterations max_retries=5; %Maximum number of retires dtmin=(1/numTimeSteps)/100; %Minimum time step size dtmax=(1/numTimeSteps); %Maximum time step size min_residual=1e-20; runMode='external';
%Specifying dimensions and number of elements r=0.5; %Radii, results in a width of 1 n=4; nCopies=n*ones(1,3); %Number of offset copies d=2*r; %Diameter w=(n-1)*d; %sampleSize
Create lattice
switch latticeType case 1 %Octet truss [Er,Vr,Cr,Fr,CFr]=rhombicDodecahedronMesh(r,nCopies); Vr=Vr./(n-1); Vr=Vr*sampleSize; [indBoundary]=tesBoundary(Fr); cPar.shrinkFactor=0.15; %Strut sides are formed by shrinking the input mesh faces by this factor cPar.meshType='hex'; %desired output mesh type cPar.indBoundary=indBoundary; %indices of the boundary faces cPar.hexSplit=2; cPar.latticeSide=2; %1=side 1 the edge lattice, 2=side 2 the dual lattice to the edge lattice [E,V,C]=element2lattice(Er,Vr,cPar); %Get lattice structure logicKeep1=~(V(:,1)<=-1e-3); logicKeep2=~(V(:,2)<=-1e-3); logicKeep3=~(V(:,3)<=-1e-3); logicKeep4=~(V(:,1)>=sampleSize+1e-3); logicKeep5=~(V(:,2)>=sampleSize+1e-3); logicKeep6=~(V(:,3)>=sampleSize+1e-3); logicKeepEs=sum(logicKeep1(E),2)>=4 &... sum(logicKeep2(E),2)>=4 &... sum(logicKeep3(E),2)>=4 &... sum(logicKeep4(E),2)>=4 &... sum(logicKeep5(E),2)>=4 &... sum(logicKeep6(E),2)>=4; E=E(logicKeepEs,:); C=C(logicKeepEs,:); [E,V,indFix]=patchCleanUnused(E,V); % [Es,Vs,~,~]=subHex(Es,Vs,1,1); % Cs=repmat(Cs,8,1); % Create patch Data for visualization [Fs,CsF]=element2patch(E,C); %Patch data for plotting %Get new boundary set indB=tesBoundary(Fs); Fb=Fs(indB,:); case 2 %Rhombic dodecahedron mesh ("dual" of octet truss lattice) [Er,Vr,Cr,Fr,CFr]=rhombicDodecahedronMesh(r,nCopies); Vr=Vr./(n-1); Vr=Vr*sampleSize; [indBoundary]=tesBoundary(Fr); cPar.shrinkFactor=0.15; %Strut sides are formed by shrinking the input mesh faces by this factor cPar.meshType='hex'; %desired output mesh type cPar.indBoundary=indBoundary; %indices of the boundary faces cPar.hexSplit=2; cPar.latticeSide=1; %1=side 1 the edge lattice, 2=side 2 the dual lattice to the edge lattice [E,V,C]=element2lattice(Er,Vr,cPar); %Get lattice structure logicKeep1=~(V(:,1)<=-1e-3); logicKeep2=~(V(:,2)<=-1e-3); logicKeep3=~(V(:,3)<=-1e-3); logicKeep4=~(V(:,1)>=sampleSize+1e-3); logicKeep5=~(V(:,2)>=sampleSize+1e-3); logicKeep6=~(V(:,3)>=sampleSize+1e-3); logicKeepEs=sum(logicKeep1(E),2)>=4 &... sum(logicKeep2(E),2)>=4 &... sum(logicKeep3(E),2)>=4 &... sum(logicKeep4(E),2)>=4 &... sum(logicKeep5(E),2)>=4 &... sum(logicKeep6(E),2)>=4; E=E(logicKeepEs,:); C=C(logicKeepEs,:); [E,V,indFix]=patchCleanUnused(E,V); % [Es,Vs,~,~]=subHex(Es,Vs,1,1); % Cs=repmat(Cs,8,1); % Create patch Data for visualization [Fs,CsF]=element2patch(E,C); %Patch data for plotting %Get new boundary set indB=tesBoundary(Fs); Fb=Fs(indB,:); case 3 boxDim=sampleSize*[1 1 1]; boxEl=[2 2 2]; [meshStruct]=hexMeshBox(boxDim,boxEl); Er=meshStruct.E; Vr=meshStruct.V; minV=min(Vr,[],1); %Get lower left front corner Vr=Vr-minV(ones(size(Vr,1),1),:); %Set corner as origin [Er,Vr,~]=hex2tet(Er,Vr,[],1); %Convert to tetrahedral elements [Fr,Cr]=element2patch(Er,[]); %Patch data for plotting [indBoundary]=tesBoundary(Fr); % Create lattice structure controlParameter.latticeSide=1; controlParameter.numDigitKeep=5; %used for merging nodes controlParameter.indBoundary=indBoundary; %indices of the boundary faces controlParameter.shrinkFactor=0.15; controlParameter.meshType='hex'; controlParameter.hexSplit=2; [E,V,C]=element2lattice(Er,Vr,controlParameter); %Get lattice structure % Create patch Data for visualization [Fs,CsF]=element2patch(E,C); %Patch data for plotting indB=tesBoundary(Fs); Fb=Fs(indB,:); end
Visualizing input mesh and lattic structures
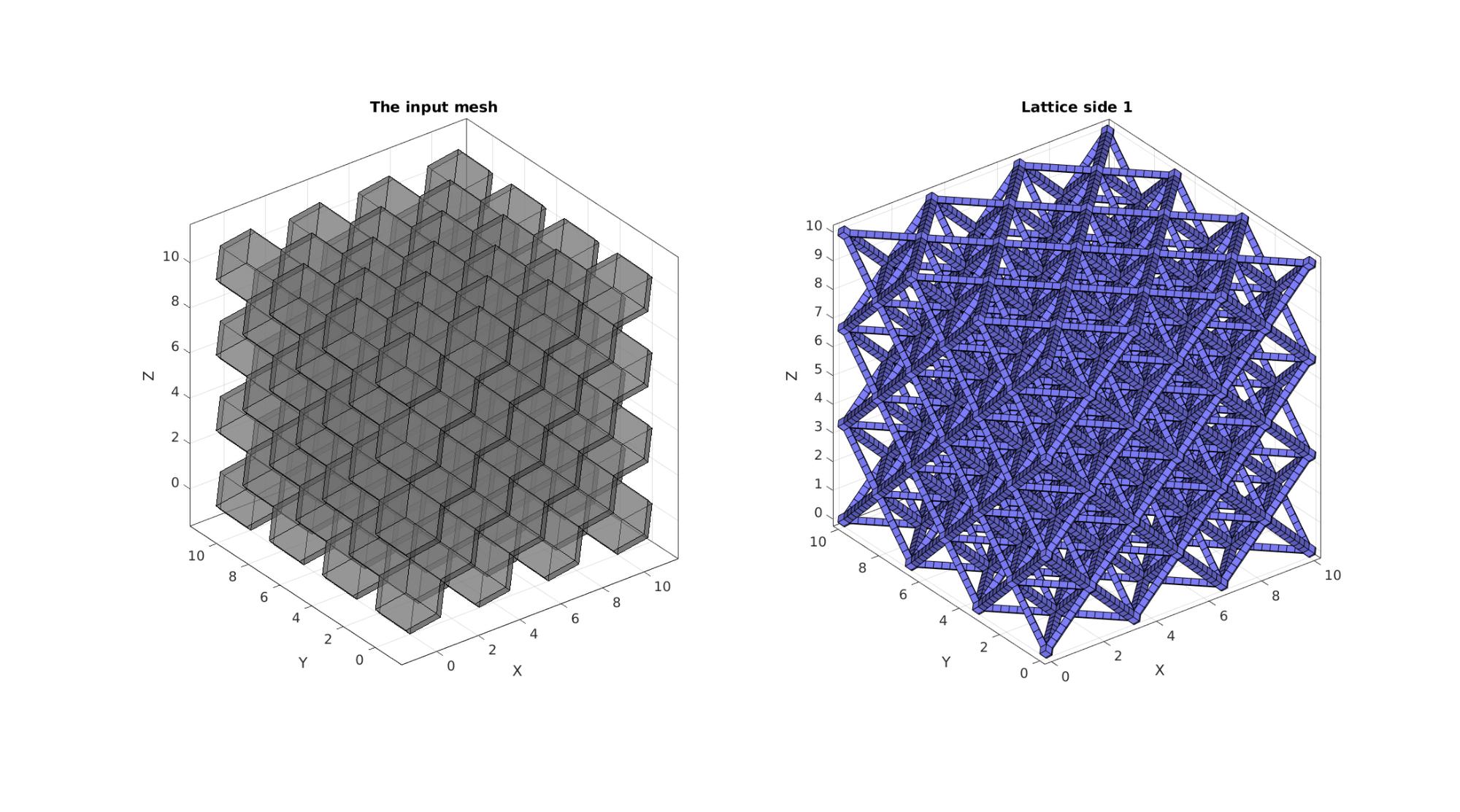
cFigure; hs=subplot(1,2,1); title('The input mesh','fontSize',fontSize) hold on; gpatch(Fr,Vr,0.5*ones(1,3),'k',0.5); axisGeom(gca,fontSize); camlight headlight; lighting flat; subplot(1,2,2); title('Lattice side 1','fontSize',fontSize) hold on; gpatch(Fb,V,'bw','k',1); % patchNormPlot(Fs,Vs); axisGeom(gca,fontSize); camlight headlight; lighting flat; drawnow;
DEFINE BC's
% Define node set logics indAll=(1:1:size(V,1))'; logicBoundary=ismember(indAll,Fb); Z=V(:,3); logicTop=Z>=(sampleSize-eps(sampleSize))& logicBoundary; logicBottom=Z<=eps(sampleSize) & logicBoundary; X=V(:,1); logicSide1=X>=(sampleSize-eps(sampleSize))& logicBoundary; logicSide2=X<=eps(sampleSize)& logicBoundary; Y=V(:,2); logicSide3=Y>=(sampleSize-eps(sampleSize))& logicBoundary; logicSide4=Y<=eps(sampleSize)& logicBoundary; %Prescribed force nodes bcPrescribeListCell{1}=find(logicSide1)'; bcPrescribeListCell{2}=find(logicSide2)'; bcPrescribeListCell{3}=find(logicSide3)'; bcPrescribeListCell{4}=find(logicSide4)'; bcPrescribeListCell{5}=find(logicTop)'; bcPrescribeListCell{6}=find(logicBottom)';
Smoothing lattice
% indKeep=unique([bcPrescribeListCell{:}]); % [Fb_clean,Vb_clean,indFix]=patchCleanUnused(Fb,Vs); % % cPar.Method='HC'; % cPar.n=6; % % cPar.RigidConstraints=indFix(indKeep); % % cPar.RigidConstraints=cPar.RigidConstraints(cPar.RigidConstraints>0); % % [Vb_clean]=tesSmooth(Fb_clean,Vb_clean,[],cPar); % ind=Fb(:); % ind=unique(ind(:)); % Vs(ind,:)=Vb_clean; % cFigure; hold on; % gpatch(Fb,Vs,'bw','k',1); % % patchNormPlot(Fs,Vs); % % plotV(Vs(indKeep,:),'k.','MarkerSize',25) % axisGeom(gca,fontSize); % camlight headlight; lighting flat; % drawnow;
Visualizing input mesh and lattic structures
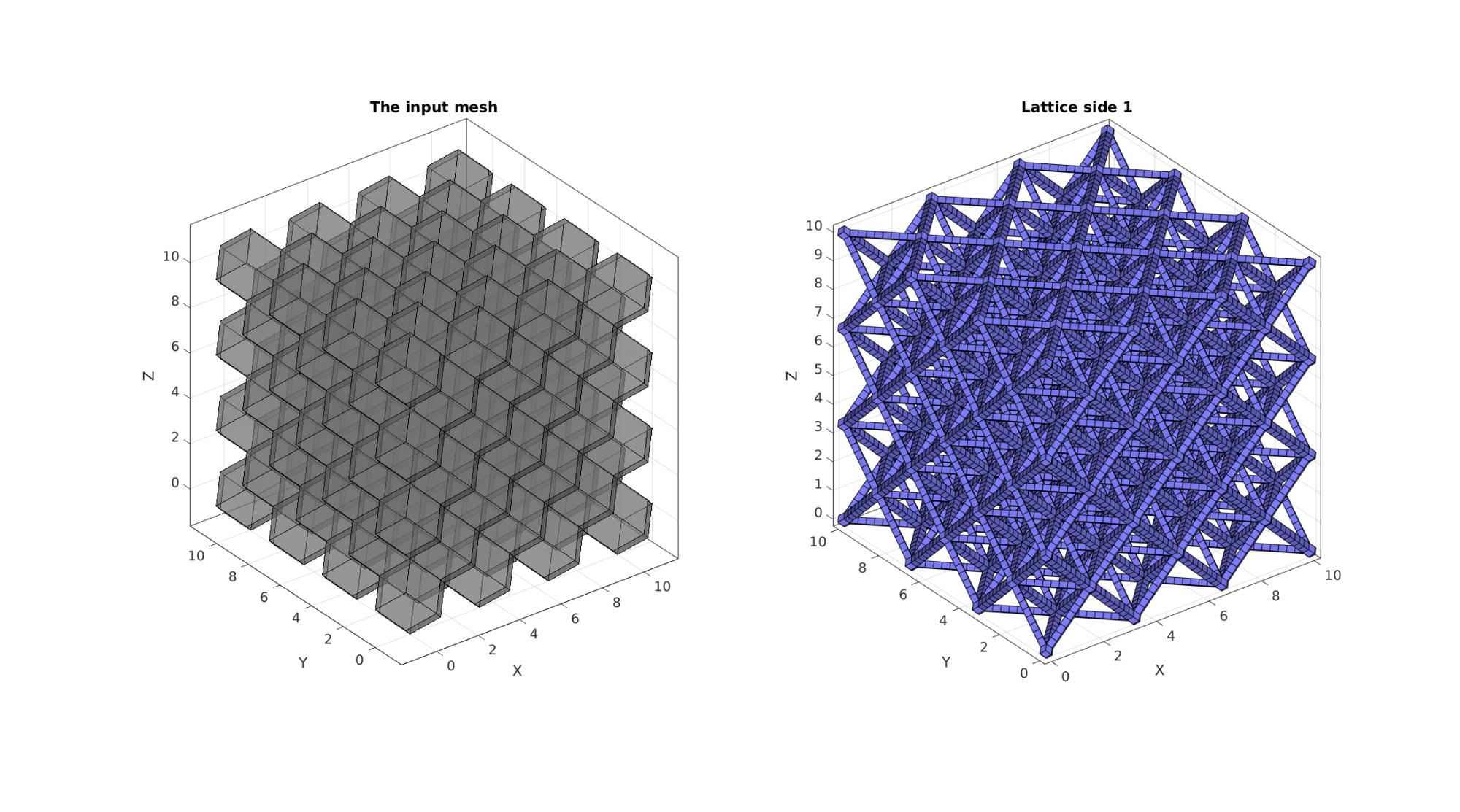
cFigure; hs=subplot(1,2,1); title('The input mesh','fontSize',fontSize) hold on; gpatch(Fr,Vr,0.5*ones(1,3),'k',0.5); axisGeom(gca,fontSize); camlight headlight; lighting flat; subplot(1,2,2); title('Lattice side 1','fontSize',fontSize) hold on; gpatch(Fb,V,'bw'); % patchNormPlot(Fs,Vs); axisGeom(gca,fontSize); camlight headlight; lighting flat; drawnow;
Visualize BC's
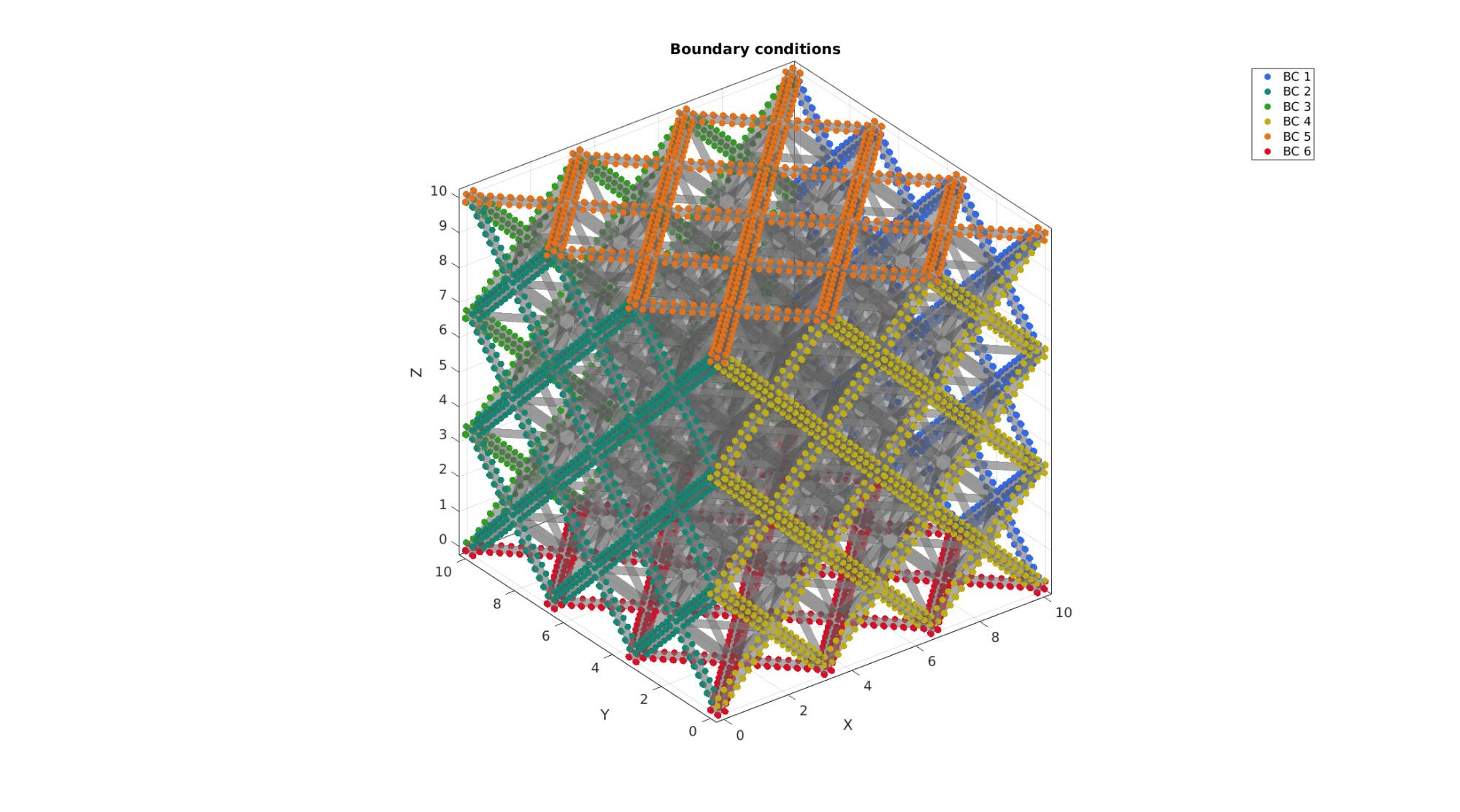
cFigure; hold on; title('Boundary conditions','FontSize',fontSize); gpatch(Fb,V,'kw','none',0.4); hl=gobjects(1,6); plotColors=gjet(6); for q=1:1:numel(bcPrescribeListCell) hl(q)=plotV(V(bcPrescribeListCell{q},:),'k.','MarkerSize',markerSize); hl(q).Color=plotColors(q,:); end legend(hl,{'BC 1','BC 2','BC 3','BC 4','BC 5','BC 6'}); axisGeom; camlight headlight; set(gca,'FontSize',fontSize); drawnow;
Defining the FEBio input structure
See also febioStructTemplate and febioStruct2xml and the FEBio user manual.
%Get a template with default settings [febio_spec]=febioStructTemplate; %febio_spec version febio_spec.ATTR.version='4.0'; %Module section febio_spec.Module.ATTR.type='solid'; %Control section febio_spec.Control.analysis='STATIC'; febio_spec.Control.time_steps=numTimeSteps; febio_spec.Control.step_size=1/numTimeSteps; febio_spec.Control.solver.max_refs=max_refs; febio_spec.Control.solver.qn_method.max_ups=max_ups; febio_spec.Control.time_stepper.dtmin=dtmin; febio_spec.Control.time_stepper.dtmax=dtmax; febio_spec.Control.time_stepper.max_retries=max_retries; febio_spec.Control.time_stepper.opt_iter=opt_iter; %Material section materialName1='Material1'; febio_spec.Material.material{1}.ATTR.name=materialName1; febio_spec.Material.material{1}.ATTR.type='Ogden'; febio_spec.Material.material{1}.ATTR.id=1; febio_spec.Material.material{1}.c1=c1; febio_spec.Material.material{1}.m1=m1; % febio_spec.Material.material{1}.c2=c1; % febio_spec.Material.material{1}.m2=-m1; febio_spec.Material.material{1}.k=k; % Mesh section % -> Nodes febio_spec.Mesh.Nodes{1}.ATTR.name='Object1_lattice'; %The node set name febio_spec.Mesh.Nodes{1}.node.ATTR.id=(1:size(V,1))'; %The node id's febio_spec.Mesh.Nodes{1}.node.VAL=V; %The nodel coordinates % -> Elements partName1='Part1_lattice'; febio_spec.Mesh.Elements{1}.ATTR.name=partName1; %Name of this part febio_spec.Mesh.Elements{1}.ATTR.type='hex8'; %Element type febio_spec.Mesh.Elements{1}.elem.ATTR.id=(1:1:size(E,1))'; %Element id's febio_spec.Mesh.Elements{1}.elem.VAL=E; %The element matrix % -> NodeSets for q=1:1:numel(bcPrescribeListCell) febio_spec.Mesh.NodeSet{q}.ATTR.name=['bcPrescribeList_',num2str(q)]; febio_spec.Mesh.NodeSet{q}.VAL=mrow(bcPrescribeListCell{q}); end %MeshDomains section febio_spec.MeshDomains.SolidDomain.ATTR.name=partName1; febio_spec.MeshDomains.SolidDomain.ATTR.mat=materialName1; %Boundary condition section % -> Prescribe boundary conditions directionStringSet={'x','x','y','y','z','z'}; displacementMagnitudeDir=[1 -1 1 -1 1 -1]; for q=1:1:numel(bcPrescribeListCell) nodeSetName=febio_spec.Mesh.NodeSet{q}.ATTR.name; febio_spec.Boundary.bc{q}.ATTR.name=['prescibed_displacement_',directionStringSet{q}]; febio_spec.Boundary.bc{q}.ATTR.type='prescribed displacement'; febio_spec.Boundary.bc{q}.ATTR.node_set=nodeSetName; febio_spec.Boundary.bc{q}.dof=directionStringSet{q}; febio_spec.Boundary.bc{q}.value.ATTR.lc=1; febio_spec.Boundary.bc{q}.value.VAL=displacementMagnitudeDir(q).*displacementMagnitude; febio_spec.Boundary.bc{q}.relative=0; end %LoadData section % -> load_controller febio_spec.LoadData.load_controller{1}.ATTR.name='LC_1'; febio_spec.LoadData.load_controller{1}.ATTR.id=1; febio_spec.LoadData.load_controller{1}.ATTR.type='loadcurve'; febio_spec.LoadData.load_controller{1}.interpolate='LINEAR'; %febio_spec.LoadData.load_controller{1}.extend='CONSTANT'; febio_spec.LoadData.load_controller{1}.points.pt.VAL=[0 0; 1 1]; %Output section % -> log file febio_spec.Output.logfile.ATTR.file=febioLogFileName; febio_spec.Output.logfile.node_data{1}.ATTR.file=febioLogFileName_disp; febio_spec.Output.logfile.node_data{1}.ATTR.data='ux;uy;uz'; febio_spec.Output.logfile.node_data{1}.ATTR.delim=','; febio_spec.Output.logfile.node_data{2}.ATTR.file=febioLogFileName_force; febio_spec.Output.logfile.node_data{2}.ATTR.data='Rx;Ry;Rz'; febio_spec.Output.logfile.node_data{2}.ATTR.delim=','; febio_spec.Output.logfile.element_data{1}.ATTR.file=febioLogFileName_stress; febio_spec.Output.logfile.element_data{1}.ATTR.data='sz'; febio_spec.Output.logfile.element_data{1}.ATTR.delim=','; % Plotfile section febio_spec.Output.plotfile.compression=0;
Quick viewing of the FEBio input file structure
The febView function can be used to view the xml structure in a MATLAB figure window.
febView(febio_spec); %Viewing the febio file
Exporting the FEBio input file
Exporting the febio_spec structure to an FEBio input file is done using the febioStruct2xml function.
febioStruct2xml(febio_spec,febioFebFileName); %Exporting to file and domNode %system(['gedit ',febioFebFileName,' &']);
Running the FEBio analysis
To run the analysis defined by the created FEBio input file the runMonitorFEBio function is used. The input for this function is a structure defining job settings e.g. the FEBio input file name. The optional output runFlag informs the user if the analysis was run succesfully.
febioAnalysis.run_filename=febioFebFileName; %The input file name febioAnalysis.run_logname=febioLogFileName; %The name for the log file febioAnalysis.disp_on=1; %Display information on the command window febioAnalysis.runMode=runMode; febioAnalysis.maxLogCheckTime=10; %Max log file checking time [runFlag]=runMonitorFEBio(febioAnalysis);%START FEBio NOW!!!!!!!!
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
--------> RUNNING/MONITORING FEBIO JOB <-------- 21-Apr-2023 11:44:07
FEBio path: /home/kevin/FEBioStudio2/bin/febio4
# Attempt removal of existing log files 21-Apr-2023 11:44:07
* Removal succesful 21-Apr-2023 11:44:07
# Attempt removal of existing .xplt files 21-Apr-2023 11:44:07
* Removal succesful 21-Apr-2023 11:44:07
# Starting FEBio... 21-Apr-2023 11:44:07
Max. total analysis time is: Inf s
* Waiting for log file creation 21-Apr-2023 11:44:07
Max. wait time: 10 s
* Log file found. 21-Apr-2023 11:44:07
# Parsing log file... 21-Apr-2023 11:44:07
number of iterations : 2 21-Apr-2023 11:44:08
number of reformations : 2 21-Apr-2023 11:44:08
------- converged at time : 0.04 21-Apr-2023 11:44:08
number of iterations : 32 21-Apr-2023 11:44:19
number of reformations : 32 21-Apr-2023 11:44:19
------- converged at time : 0.08 21-Apr-2023 11:44:19
number of iterations : 11 21-Apr-2023 11:44:23
number of reformations : 11 21-Apr-2023 11:44:23
------- converged at time : 0.0975473 21-Apr-2023 11:44:23
number of iterations : 11 21-Apr-2023 11:44:27
number of reformations : 11 21-Apr-2023 11:44:27
------- converged at time : 0.110611 21-Apr-2023 11:44:27
number of iterations : 8 21-Apr-2023 11:44:31
number of reformations : 8 21-Apr-2023 11:44:31
------- converged at time : 0.118739 21-Apr-2023 11:44:31
number of iterations : 6 21-Apr-2023 11:44:34
number of reformations : 6 21-Apr-2023 11:44:34
------- converged at time : 0.125831 21-Apr-2023 11:44:34
number of iterations : 5 21-Apr-2023 11:44:35
number of reformations : 5 21-Apr-2023 11:44:35
------- converged at time : 0.132924 21-Apr-2023 11:44:35
number of iterations : 4 21-Apr-2023 11:44:37
number of reformations : 4 21-Apr-2023 11:44:37
------- converged at time : 0.143157 21-Apr-2023 11:44:37
number of iterations : 4 21-Apr-2023 11:44:38
number of reformations : 4 21-Apr-2023 11:44:38
------- converged at time : 0.159343 21-Apr-2023 11:44:38
number of iterations : 4 21-Apr-2023 11:44:40
number of reformations : 4 21-Apr-2023 11:44:40
------- converged at time : 0.180292 21-Apr-2023 11:44:40
number of iterations : 4 21-Apr-2023 11:44:42
number of reformations : 4 21-Apr-2023 11:44:42
------- converged at time : 0.205052 21-Apr-2023 11:44:42
number of iterations : 8 21-Apr-2023 11:44:45
number of reformations : 8 21-Apr-2023 11:44:45
------- converged at time : 0.232859 21-Apr-2023 11:44:45
number of iterations : 6 21-Apr-2023 11:44:47
number of reformations : 6 21-Apr-2023 11:44:47
------- converged at time : 0.256995 21-Apr-2023 11:44:47
number of iterations : 5 21-Apr-2023 11:44:49
number of reformations : 5 21-Apr-2023 11:44:49
------- converged at time : 0.28113 21-Apr-2023 11:44:49
number of iterations : 9 21-Apr-2023 11:44:52
number of reformations : 9 21-Apr-2023 11:44:52
------- converged at time : 0.30678 21-Apr-2023 11:44:52
number of iterations : 4 21-Apr-2023 11:44:54
number of reformations : 4 21-Apr-2023 11:44:54
------- converged at time : 0.327797 21-Apr-2023 11:44:54
number of iterations : 4 21-Apr-2023 11:44:55
number of reformations : 4 21-Apr-2023 11:44:55
------- converged at time : 0.35261 21-Apr-2023 11:44:55
number of iterations : 4 21-Apr-2023 11:44:57
number of reformations : 4 21-Apr-2023 11:44:57
------- converged at time : 0.38046 21-Apr-2023 11:44:57
number of iterations : 4 21-Apr-2023 11:44:58
number of reformations : 4 21-Apr-2023 11:44:58
------- converged at time : 0.41074 21-Apr-2023 11:44:58
number of iterations : 4 21-Apr-2023 11:45:00
number of reformations : 4 21-Apr-2023 11:45:00
------- converged at time : 0.442965 21-Apr-2023 11:45:00
number of iterations : 4 21-Apr-2023 11:45:02
number of reformations : 4 21-Apr-2023 11:45:02
------- converged at time : 0.476744 21-Apr-2023 11:45:02
number of iterations : 4 21-Apr-2023 11:45:03
number of reformations : 4 21-Apr-2023 11:45:03
------- converged at time : 0.511768 21-Apr-2023 11:45:03
number of iterations : 4 21-Apr-2023 11:45:05
number of reformations : 4 21-Apr-2023 11:45:05
------- converged at time : 0.547787 21-Apr-2023 11:45:05
number of iterations : 4 21-Apr-2023 11:45:07
number of reformations : 4 21-Apr-2023 11:45:07
------- converged at time : 0.584602 21-Apr-2023 11:45:07
number of iterations : 4 21-Apr-2023 11:45:08
number of reformations : 4 21-Apr-2023 11:45:08
------- converged at time : 0.622054 21-Apr-2023 11:45:08
number of iterations : 4 21-Apr-2023 11:45:10
number of reformations : 4 21-Apr-2023 11:45:10
------- converged at time : 0.660015 21-Apr-2023 11:45:10
number of iterations : 4 21-Apr-2023 11:45:11
number of reformations : 4 21-Apr-2023 11:45:11
------- converged at time : 0.698385 21-Apr-2023 11:45:11
number of iterations : 4 21-Apr-2023 11:45:13
number of reformations : 4 21-Apr-2023 11:45:13
------- converged at time : 0.73708 21-Apr-2023 11:45:13
number of reformations : 4 21-Apr-2023 11:45:15
------- converged at time : 0.776037 21-Apr-2023 11:45:15
number of iterations : 4 21-Apr-2023 11:45:16
number of reformations : 4 21-Apr-2023 11:45:16
------- converged at time : 0.815202 21-Apr-2023 11:45:16
number of iterations : 4 21-Apr-2023 11:45:18
number of reformations : 4 21-Apr-2023 11:45:18
------- converged at time : 0.854534 21-Apr-2023 11:45:18
number of iterations : 4 21-Apr-2023 11:45:19
number of reformations : 4 21-Apr-2023 11:45:19
------- converged at time : 0.893999 21-Apr-2023 11:45:19
number of iterations : 4 21-Apr-2023 11:45:21
number of reformations : 4 21-Apr-2023 11:45:21
------- converged at time : 0.933572 21-Apr-2023 11:45:21
number of iterations : 4 21-Apr-2023 11:45:22
number of reformations : 4 21-Apr-2023 11:45:22
------- converged at time : 0.97323 21-Apr-2023 11:45:22
number of iterations : 3 21-Apr-2023 11:45:24
number of reformations : 3 21-Apr-2023 11:45:24
------- converged at time : 1 21-Apr-2023 11:45:24
Elapsed time : 0:01:17 21-Apr-2023 11:45:24
N O R M A L T E R M I N A T I O N
# Done 21-Apr-2023 11:45:24
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
Import FEBio results
if runFlag==1 %i.e. a succesful run
% Importing nodal displacements from a log file dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_disp),0,1); %Access data N_disp_mat=dataStruct.data; %Displacement timeVec=dataStruct.time; %Time %Create deformed coordinate set V_DEF=N_disp_mat+repmat(V,[1 1 size(N_disp_mat,3)]);
% Importing nodal forces
dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_force),0,1);
N_force_mat=dataStruct.data;
indicesSide=bcPrescribeListCell{1};
areaSide=sampleSize.^2;
stressVal=mean(squeeze(N_force_mat(indicesSide,1,:))./areaSide,1);
J_Val=1-((1-J_final).*timeVec(:));
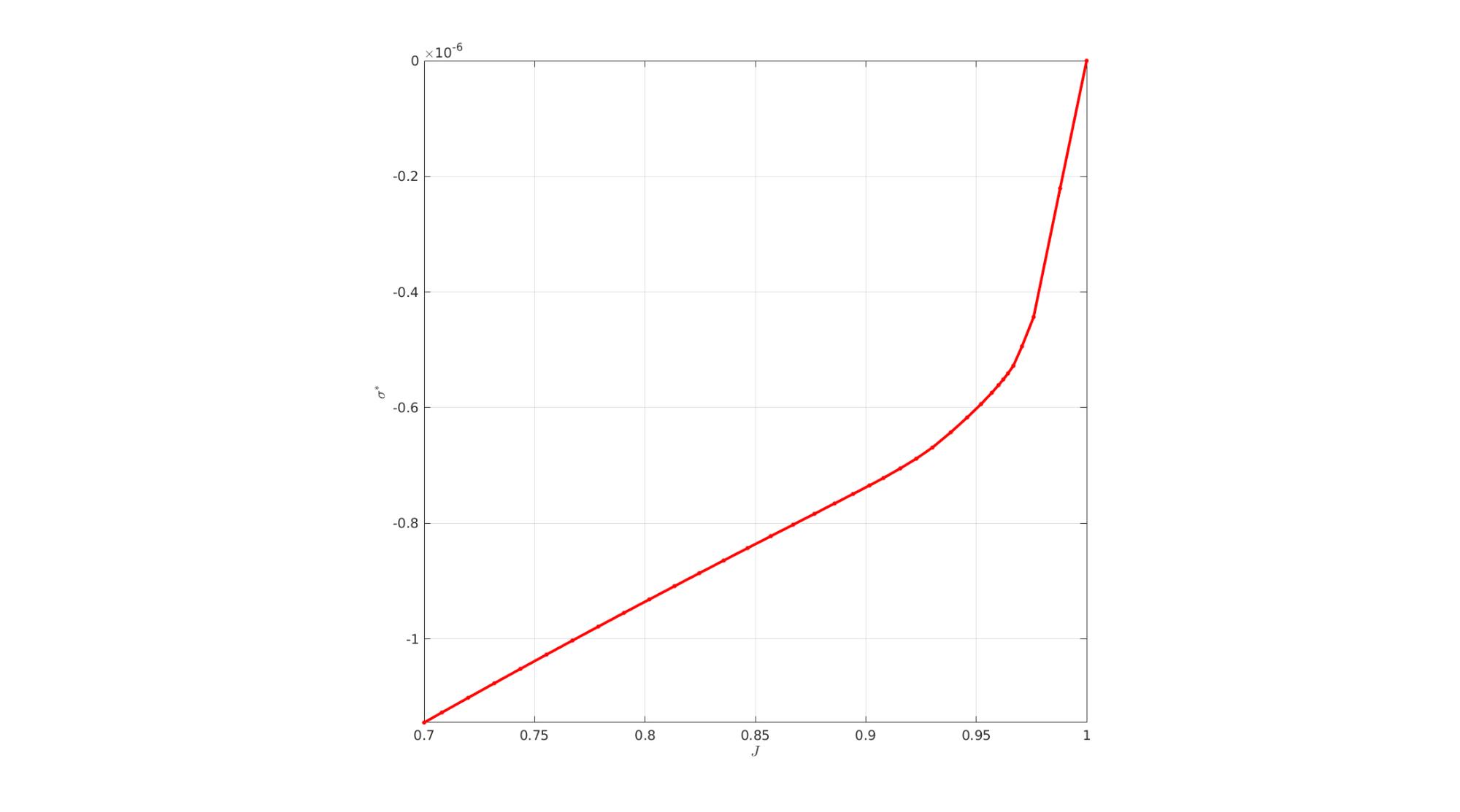
cFigure; hold on; xlabel('$J$','Interpreter','Latex'); ylabel('$\sigma^*$','Interpreter','Latex'); plot(J_Val(:),stressVal(:),'r.-','LineWidth',3,'MarkerSize',15); axis square; axis tight; grid on; box on; set(gca,'FontSize',fontSize); drawnow;
Plotting the simulated results using anim8 to visualize and animate deformations
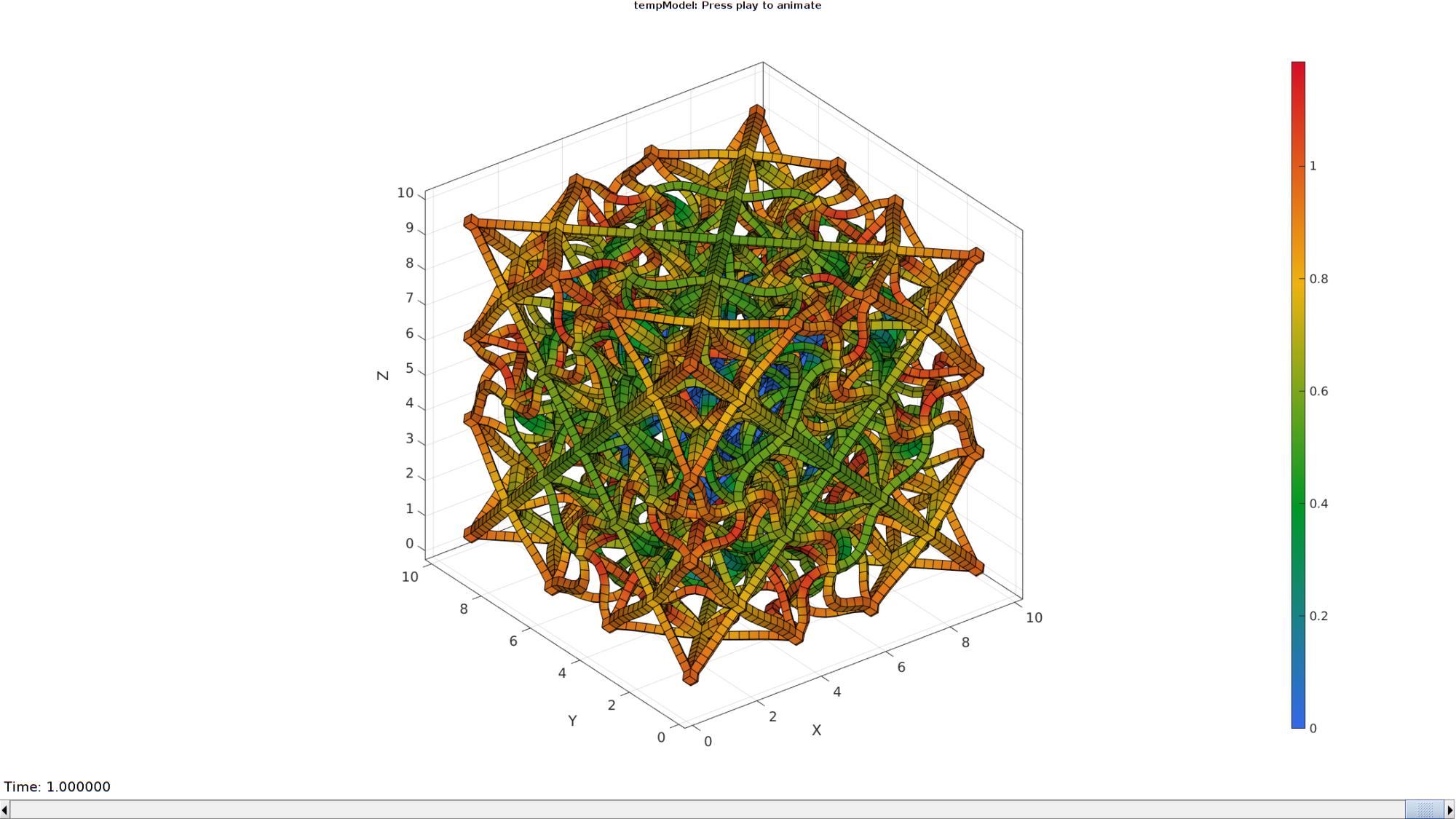
DN_magnitude=sqrt(sum(N_disp_mat(:,:,end).^2,2)); %Current displacement magnitude % Create basic view and store graphics handle to initiate animation hf=cFigure; %Open figure gtitle([febioFebFileNamePart,': Press play to animate']); hp=gpatch(Fb,V_DEF(:,:,end),DN_magnitude,'k',1); %Add graphics object to animate % gpatch(Fb,Vs,'kw','none',0.25); %A static graphics object hp.FaceColor='interp'; axisGeom(gca,fontSize); colormap(gjet(250)); colorbar; caxis([0 max(DN_magnitude)]); axis(axisLim(V_DEF)); %Set axis limits statically % view(130,25); %Set view direction camlight headlight; % Set up animation features animStruct.Time=timeVec; %The time vector for qt=1:1:size(N_disp_mat,3) %Loop over time increments DN=N_disp_mat(:,:,qt); %Current displacement DN_magnitude=sqrt(sum(DN.^2,2)); %Current displacement magnitude %Set entries in animation structure animStruct.Handles{qt}=[hp hp]; %Handles of objects to animate animStruct.Props{qt}={'Vertices','CData'}; %Properties of objects to animate animStruct.Set{qt}={V_DEF(:,:,qt),DN_magnitude}; %Property values for to set in order to animate end anim8(hf,animStruct); %Initiate animation feature drawnow;
end

GIBBON www.gibboncode.org
Kevin Mattheus Moerman, [email protected]
GIBBON footer text
License: https://github.com/gibbonCode/GIBBON/blob/master/LICENSE
GIBBON: The Geometry and Image-based Bioengineering add-On. A toolbox for image segmentation, image-based modeling, meshing, and finite element analysis.
Copyright (C) 2006-2022 Kevin Mattheus Moerman and the GIBBON contributors
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.