DEMO_febio_0050_foot_insole_01
Below is a demonstration for:
- Building triangulated surface geometry for a foot
- Meshing the foot using tetrahedral elements
- Building surface model of an insole and meshing it with quads
- Extruding the surface to a thickened layer of hexahedral elements
- Defining the boundary conditions
- Coding the febio structure
- Running the model
- Importing and visualizing results
Contents
- Keywords
- Control parameters
- Import surface models
- Subdivide skin mesh and smooth
- Compute mesh derived parameters
- Reorient surfaces
- Cut skin surface
- Scale foot to desired size
- Close over top of skin
- Joining surface features
- Merge shared nodes
- Mesh foot with tetrahedral elements
- Build insole
- Joining node sets
- Define contact surfaces
- Define boundary conditions
- Set-up materials
- Convert list of materials to element id
- Defining the FEBio input structure
- Quick viewing of the FEBio input file structure
- Running the FEBio analysis
- Exporting the FEBio input file
- Run febio
- Set displacement magnitude in input file structure
- Export input file
- Run febio
- Importing rigid body reaction forces from a log file
- Use inter/extra-polation to guess displacement
- Import rigid body reaction forces
- Visualize force
- Plot animation
Keywords
- febio_spec version 4.0
- febio, FEBio
- shoe insole
- contact, sliding, friction
- tetrahedral elements, tet4
- hexahedral elements, hex8
- static, solid
- hyperelastic, Ogden
- displacement logfile
clear; close all; clc;
Plot settings
fontSize=15; faceAlpha1=1; faceAlpha2=0.3; markerSize1=15; markerSize2=10; lineWidth=2;
Control parameters
% Path names defaultFolder = fileparts(fileparts(mfilename('fullpath'))); savePath=fullfile(defaultFolder,'data','temp'); loadPathSurfaces=fullfile(defaultFolder,'data','STL','leg','post'); % Defining file names febioFebFileNamePart='tempModel'; febioFebFileName=fullfile(savePath,[febioFebFileNamePart,'.feb']); %FEB file name febioLogFileName=fullfile(savePath,[febioFebFileNamePart,'.txt']); %FEBio log file name febioLogFileName_disp=[febioFebFileNamePart,'_disp_out.txt']; %Log file name for exporting displacement febioLogFileName_force=[febioFebFileNamePart,'_force_out.txt']; %Log file name for exporting force febioLogFileName_sed=[febioFebFileNamePart,'_energy_out.txt']; %Log file name for exporting strain energy density % Surface model file names fileNameBones=fullfile(loadPathSurfaces,'Foot_bulk.stl'); fileNameSkin=fullfile(loadPathSurfaces,'Skin_coarse.stl'); %Geometric parameters soleOffsetOutward=3; %How much the insole protrudes outward from the foot numSmoothIterations_sole_Z=15; %Number of smoothing iterations for the sole Z value soleMinThickness=5; %Sole thickness soleMaxThickness=10; %Sole thickness volumeFactor=1; %Volume factor used in tetgen, larger means larger internal elements footSize=251; %Foot size in mm size 6=241 mm, size 7=251 mm % Material parameters %-> Tissue (Ogden, uncoupled) % Data source: Petre et al. 2013, Optimization of Nonlinear Hyperelastic % Coefficients for Foot Tissues Using a Magnetic Resonance Imaging % Deformation Experiment. https://dx.doi.org/10.1115%2F1.4023695 c1_1=0.02652; %Shear-modulus-like parameter in MPa m1_1=17.71; %Material parameter setting degree of non-linearity k_1=c1_1*100; %Bulk modulus %-> Sole %Sole material parameters (Ogden unconstrained, with m=2 i.e. Neo-Hookean) E_youngs_min=0.05; %Shear-modulus-like parameter in MPa E_youngs_max=0.1; %Shear-modulus-like parameter in MPa numMaterialLevels=50; %Max number of materials to use initialMaterialLevel=numMaterialLevels; E_youngs_range=linspace(E_youngs_min,E_youngs_max,numMaterialLevels); %Shear-modulus-like parameter nu=0.4; testCase=1; % Change to test different sole cases, e.g. stiff, soft, spatially varying. maxLevelColorbar_SED=0.0005; % FEA control settings numTimeSteps=10; %Number of time steps desired max_refs=25; %Max reforms max_ups=0; %Set to zero to use full-Newton iterations opt_iter=10; %Optimum number of iterations max_retries=8; %Maximum number of retires dtmin=(1/numTimeSteps)/100; %Minimum time step size dtmax=1/numTimeSteps; %Maximum time step size symmetric_stiffness=0; min_residual=1e-20; runMode='external'; %'internal'; initialSoleSpacing=0.1; %Boundary condition parameters displacementMagnitude=-2.63; %Displacement applied to the bones bodyWeight=65/2;% Body weight in Kg forceBody=bodyWeight.*-9.81; %Force due to body weight forceDifferenceToleranceFraction=0.05; forceDifferenceTolerance=abs(forceBody.*forceDifferenceToleranceFraction); optimizeForceOption=0; %set to 0 to turn off and use input displacement maxProposedDisplacementDifference=1; %Contact parameters contactPenalty=10; laugon=0; minaug=1; maxaug=10; fric_coeff=0.25;
Import surface models
%Import STL file for the bones [stlStruct] = import_STL(fileNameBones); F1=stlStruct.solidFaces{1}; %Faces V1=stlStruct.solidVertices{1}; %Vertices [F1,V1]=mergeVertices(F1,V1); %Merge nodes %Import STL file for the skin [stlStruct] = import_STL(fileNameSkin); F2=stlStruct.solidFaces{1}; %Faces V2=stlStruct.solidVertices{1}; %Vertices [F2,V2]=mergeVertices(F2,V2); %Merge nodes V2=V2*100; %Scale
Subdivide skin mesh and smooth
% %Split elements once % [F2,V2]=subtri(F2,V2,1); % % %Smooth % clear cParSmooth; % cParSmooth.n=5; % cParSmooth.Method='HC'; % [V2]=patchSmooth(F2,V2,[],cParSmooth);
Visualize imported surfaces
cFigure; hold on; gpatch(F1,V1,'bw','none',1); gpatch(F2,V2,'rw','none',0.5); camlight('headlight'); axisGeom(gca,fontSize); drawnow;

Compute mesh derived parameters
Compute point spacings for the surfaces meshes. These are useful to relate other mesh sizes to.
pointSpacing1=mean(patchEdgeLengths(F1,V1)); pointSpacing2=mean(patchEdgeLengths(F2,V2)); pointSpacing=mean([pointSpacing1 pointSpacing2]); pointSpacingSole=pointSpacing;
Reorient surfaces
Reorient so that the leg points up in a forward view with the toes pointing towards the viewer.
R=euler2DCM([-0.5*pi 0 -0.5*pi]); V1=V1*R; V2=V2*R;
Visualize reoriented surfaces
cFigure; hold on; gpatch(F1,V1,'bw','none',1); gpatch(F2,V2,'rw','none',0.5); camlight('headlight'); axisGeom(gca,fontSize); drawnow;
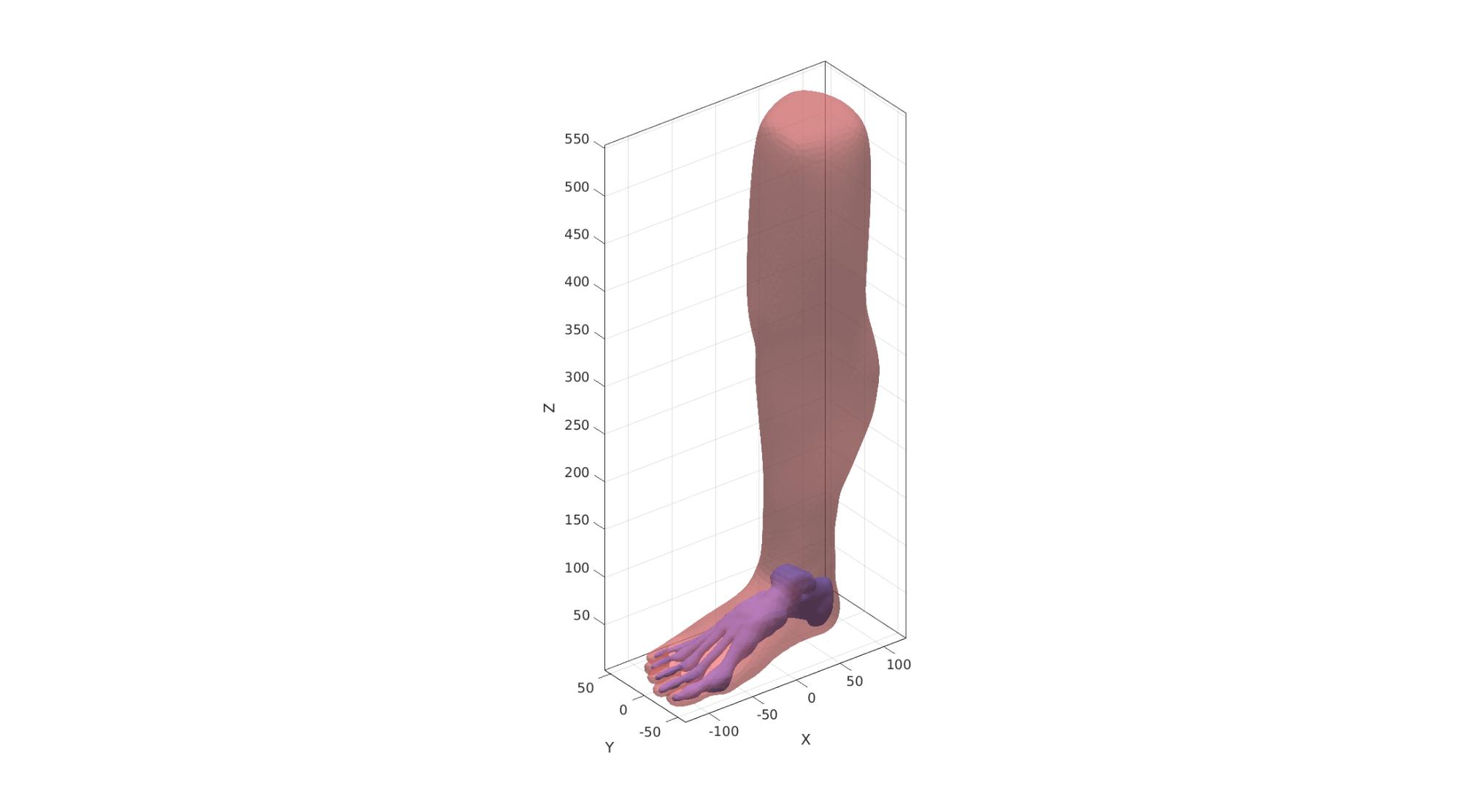
Cut skin surface
The skin surface is cut such that it stops where the bones of the ankle end in the z direction.
%Create a logic for cutting away faces max_Z1=max(V1(:,3))+2*pointSpacing; %Max Z-level used for cutting logicVertices=V2(:,3)<max_Z1; %Logic for the points below this level logicFaces=all(logicVertices(F2),2); %Logic for the faces logicFaces=triSurfLogicSharpFix(F2,logicFaces,3); %Altered logic so it is smoother
Visualize
cFigure; hold on; gpatch(F1,V1,'bw','none',1); gpatch(F2,V2,logicFaces,'none',0.5); camlight('headlight'); axisGeom(gca,fontSize); drawnow;
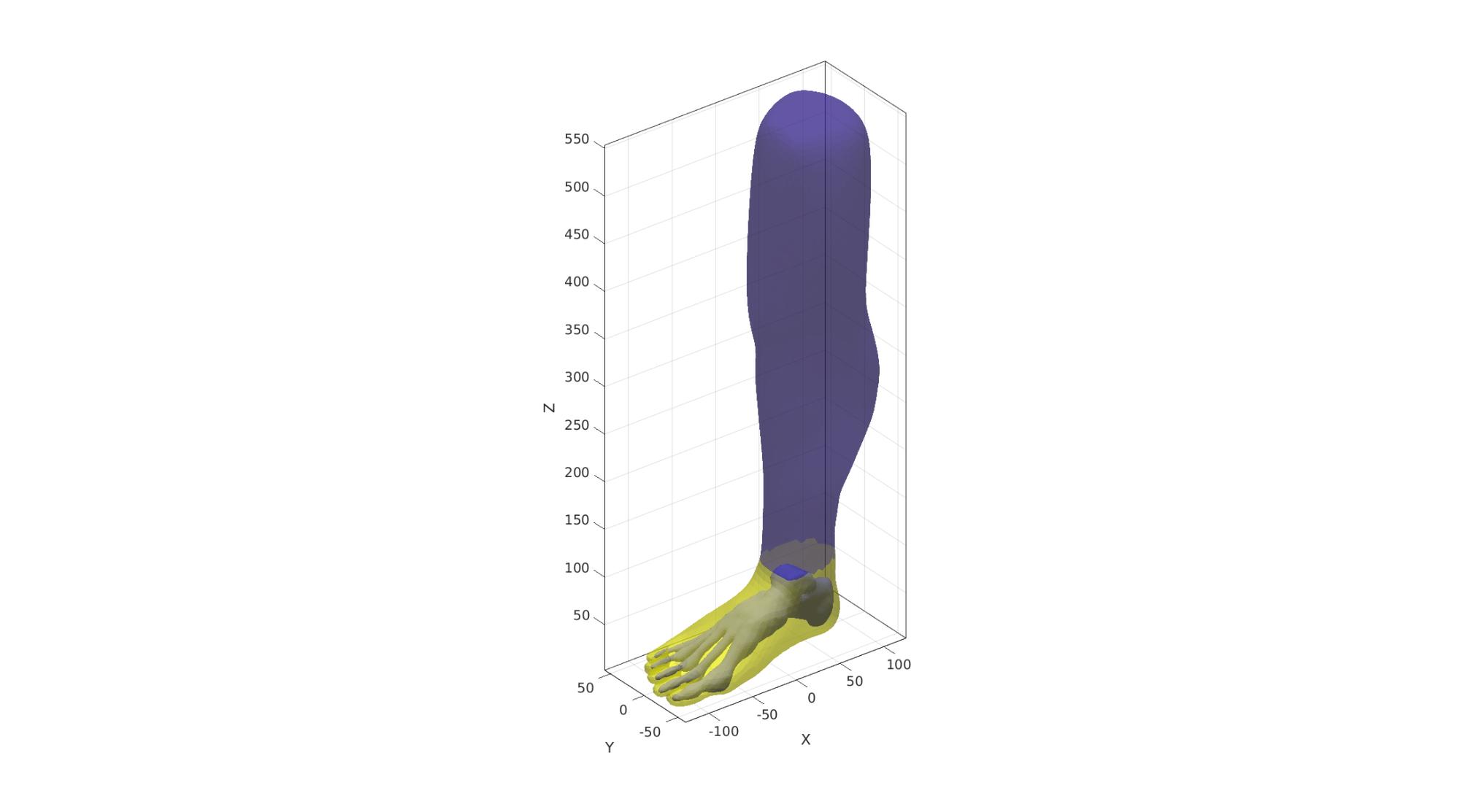
Cut away faces using logic
F2=F2(logicFaces,:); %The faces to keep [F2,V2]=patchCleanUnused(F2,V2); %Remove unused points %Attempt to self triangulate potentially jagged edge Eb=patchBoundary(F2); %Get boundary edges indBoundary=edgeListToCurve(Eb); %Convert boundary edges to a curve list indBoundary=indBoundary(1:end-1); %Trim off last point since it is equal to first on a closed loop angleThreshold=pi*(120/180); %threshold for self triangulation [F2,V2,indBoundaryTop]=triSurfSelfTriangulateBoundary(F2,V2,indBoundary,angleThreshold,1); %Force boundary to have the max Z level chosen V2(indBoundaryTop,3)=max_Z1;
Visualize
cFigure; hold on; gpatch(F1,V1,'bw','none',1); gpatch(F2,V2,'rw','k',0.5); plotV(V2(indBoundaryTop,:),'r-','LineWidth',lineWidth); camlight('headlight'); axisGeom(gca,fontSize); drawnow;
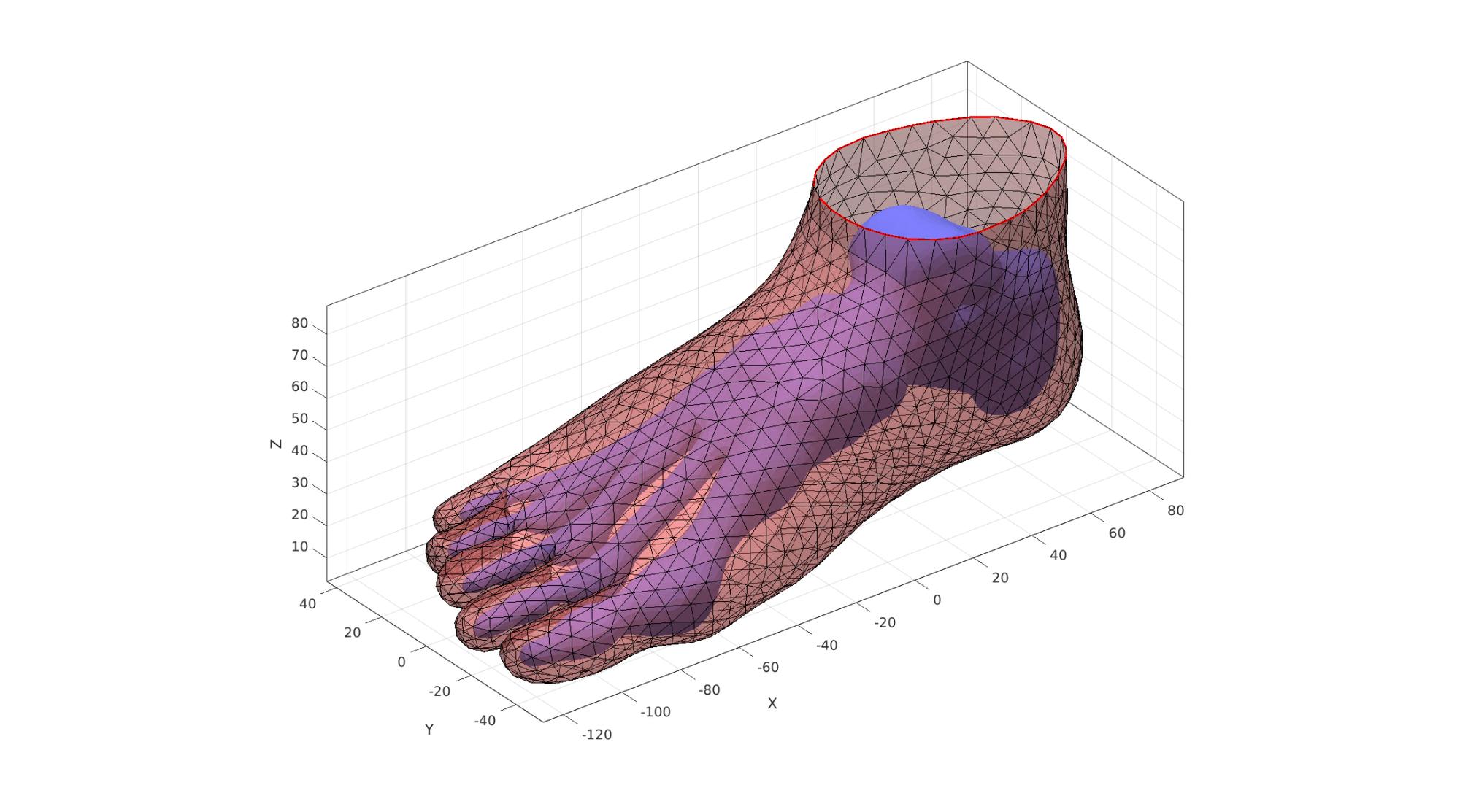
Scale foot to desired size
currentFootSize=abs(max(V2(:,1))-min(V2(:,1))); V2=V2./currentFootSize; %Scale to unit foot V2=V2.*footSize; %Scale to desired foot size V1=V1./currentFootSize; %Scale to unit foot V1=V1.*footSize; %Scale to desired foot size
Close over top of skin
The top boundary curve of the cut surface is filled with triangles. This is a 2D method. The z-coordinate is added after.
[F2t,V2t]=regionTriMesh2D({V2(indBoundaryTop,[1 2])},pointSpacing2,0,0);
V2t(:,3)=mean(V2(indBoundaryTop,3)); %Add/set z-level
Visualize
cFigure; hold on; gpatch(F1,V1,'bw','none',1); gpatch(F2,V2,'rw','k',0.5); gpatch(F2t,V2t,'gw','k',1); plotV(V2(indBoundaryTop,:),'r-','LineWidth',lineWidth); camlight('headlight'); axisGeom(gca,fontSize); drawnow;
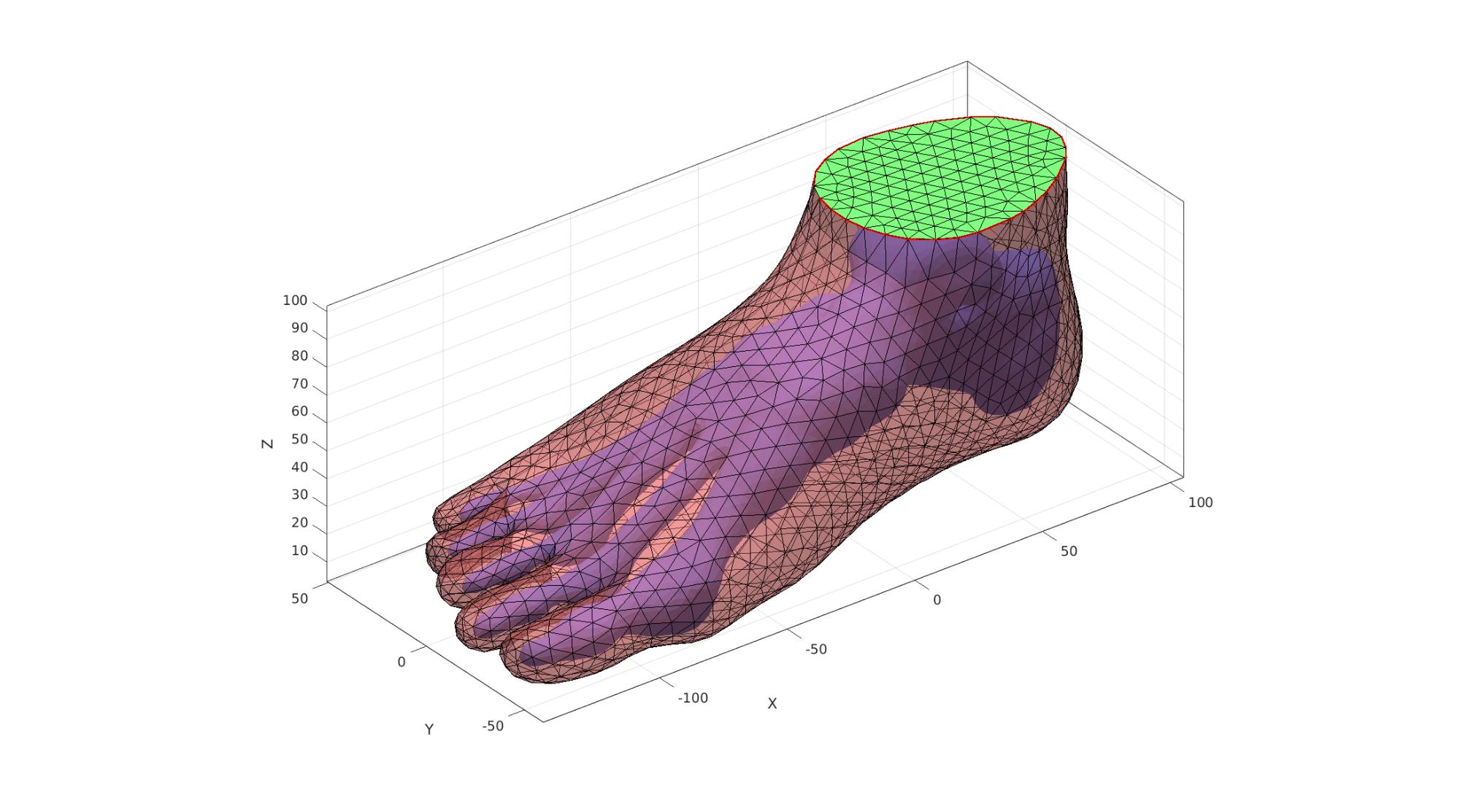
Joining surface features
Add all surface sets together in joint list of faces, vertices
[FT,VT,CT]=joinElementSets({F1,F2,F2t},{V1,V2,V2t});
Merge shared nodes
The join operation only adds the sets together. Nodes with the same coordinates are not seen as the same yet and need to be merged.
[FT,VT]=mergeVertices(FT,VT); %Merge nodes
Visualize
cFigure; hold on; gpatch(FT,VT,CT,'k',0.5); patchNormPlot(FT,VT); camlight('headlight'); axisGeom(gca,fontSize); colormap gjet; icolorbar; drawnow;
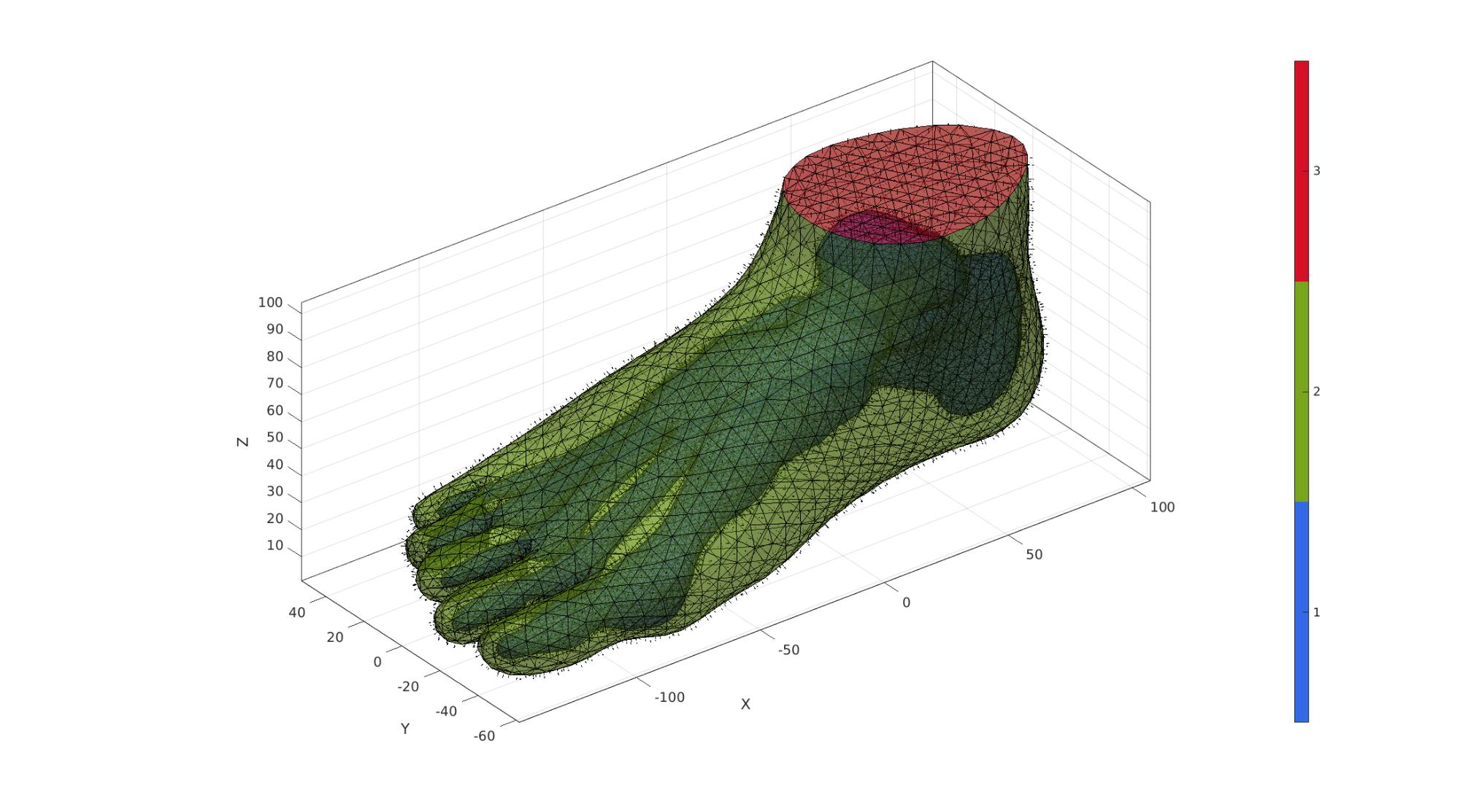
Mesh foot with tetrahedral elements
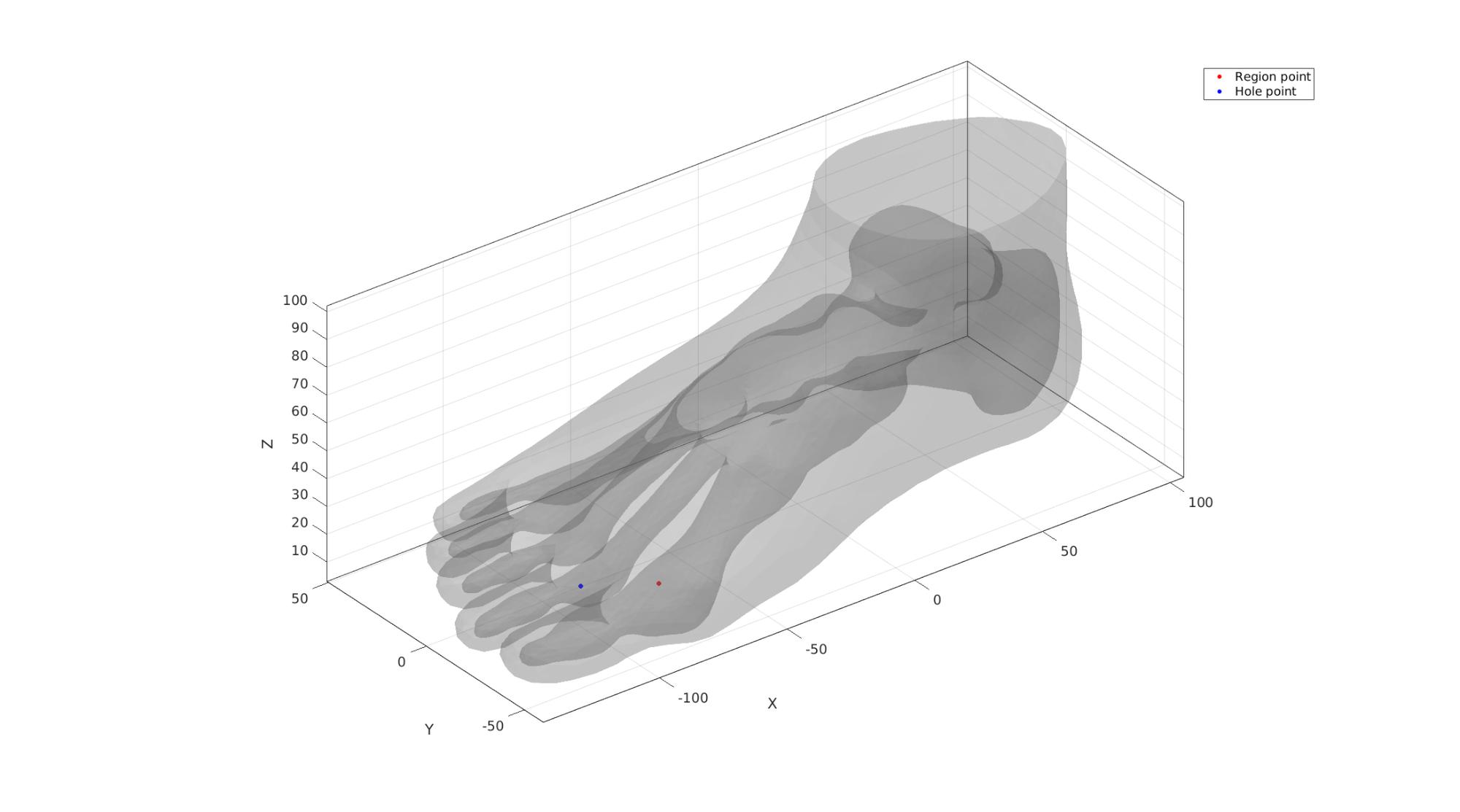
Tet meshing is based on tetgen. TetGen requires a interior points for regions to be meshed, as well as intertior points for holes.
Define region points
[V_region]=getInnerPoint({FT(CT==2 | CT==3,:),FT(CT==1,:)},{VT,VT});
[V_hole]=getInnerPoint(FT(CT==1,:),VT);
Visualize interior points
cFigure; hold on; gpatch(FT,VT,'kw','none',0.2); hp1=plotV(V_region,'r.','markerSize',markerSize1); hp2=plotV(V_hole,'b.','markerSize',markerSize1); legend([hp1 hp2],{'Region point','Hole point'}); camlight('headlight'); axisGeom(gca,fontSize); drawnow;
Mesh using tetgen
inputStruct.stringOpt='-pq1.2AaY'; %TetGen option string inputStruct.Faces=FT; %The faces inputStruct.Nodes=VT; %The vertices inputStruct.holePoints=V_hole; %The hole interior points inputStruct.faceBoundaryMarker=CT; %Face boundary markers inputStruct.regionPoints=V_region; %The region interior points inputStruct.regionA=tetVolMeanEst(F2,V2)*volumeFactor; %Volume for regular tets
Mesh model using tetrahedral elements using tetGen
[meshOutput]=runTetGen(inputStruct); %Run tetGen
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- TETGEN Tetrahedral meshing --- 27-Apr-2023 16:18:02 %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- Writing SMESH file --- 27-Apr-2023 16:18:02 ----> Adding node field ----> Adding facet field ----> Adding holes specification ----> Adding region specification --- Done --- 27-Apr-2023 16:18:02 --- Running TetGen to mesh input boundary--- 27-Apr-2023 16:18:02 Opening /mnt/data/MATLAB/GIBBON/data/temp/temp.smesh. Delaunizing vertices... Delaunay seconds: 0.031735 Creating surface mesh ... Surface mesh seconds: 0.012355 Recovering boundaries... Boundary recovery seconds: 0.025781 Removing exterior tetrahedra ... Spreading region attributes. Exterior tets removal seconds: 0.01477 Recovering Delaunayness... Delaunay recovery seconds: 0.011781 Refining mesh... 8715 insertions, added 4033 points, 82895 tetrahedra in queue. 2902 insertions, added 229 points, 0 tetrahedra in queue. Refinement seconds: 0.189777 Smoothing vertices... Mesh smoothing seconds: 0.233285 Improving mesh... Mesh improvement seconds: 0.010192 Writing /mnt/data/MATLAB/GIBBON/data/temp/temp.1.node. Writing /mnt/data/MATLAB/GIBBON/data/temp/temp.1.ele. Writing /mnt/data/MATLAB/GIBBON/data/temp/temp.1.face. Writing /mnt/data/MATLAB/GIBBON/data/temp/temp.1.edge. Output seconds: 0.122931 Total running seconds: 0.65311 Statistics: Input points: 6538 Input facets: 13068 Input segments: 19602 Input holes: 1 Input regions: 1 Mesh points: 10942 Mesh tetrahedra: 47679 Mesh faces: 101892 Mesh faces on exterior boundary: 13068 Mesh faces on input facets: 13068 Mesh edges on input segments: 19602 Steiner points inside domain: 4404 --- Done --- 27-Apr-2023 16:18:03 %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% --- Importing TetGen files --- 27-Apr-2023 16:18:03 --- Done --- 27-Apr-2023 16:18:03
Access model element and patch data
Fb_foot=meshOutput.facesBoundary; %Boundary faces of the foot Cb_foot=meshOutput.boundaryMarker; %Boundary marker/color data for the foot V_foot=meshOutput.nodes; %The vertices/nodes E_foot=meshOutput.elements; %The tet4 elements
Visualizing mesh using meshView, see also anim8
meshView(meshOutput);

Build insole
The insole is created by taking the 2D convex hull of the foot (ignoring the z-direction). Next this convex hull is resampled and filled with triangular elements. The z-coordinates are then based on the nearest foot nodes. The z-coordinate data is next smoothed to create a smooth surface. This surface is next thickened to form hexahedral elements.
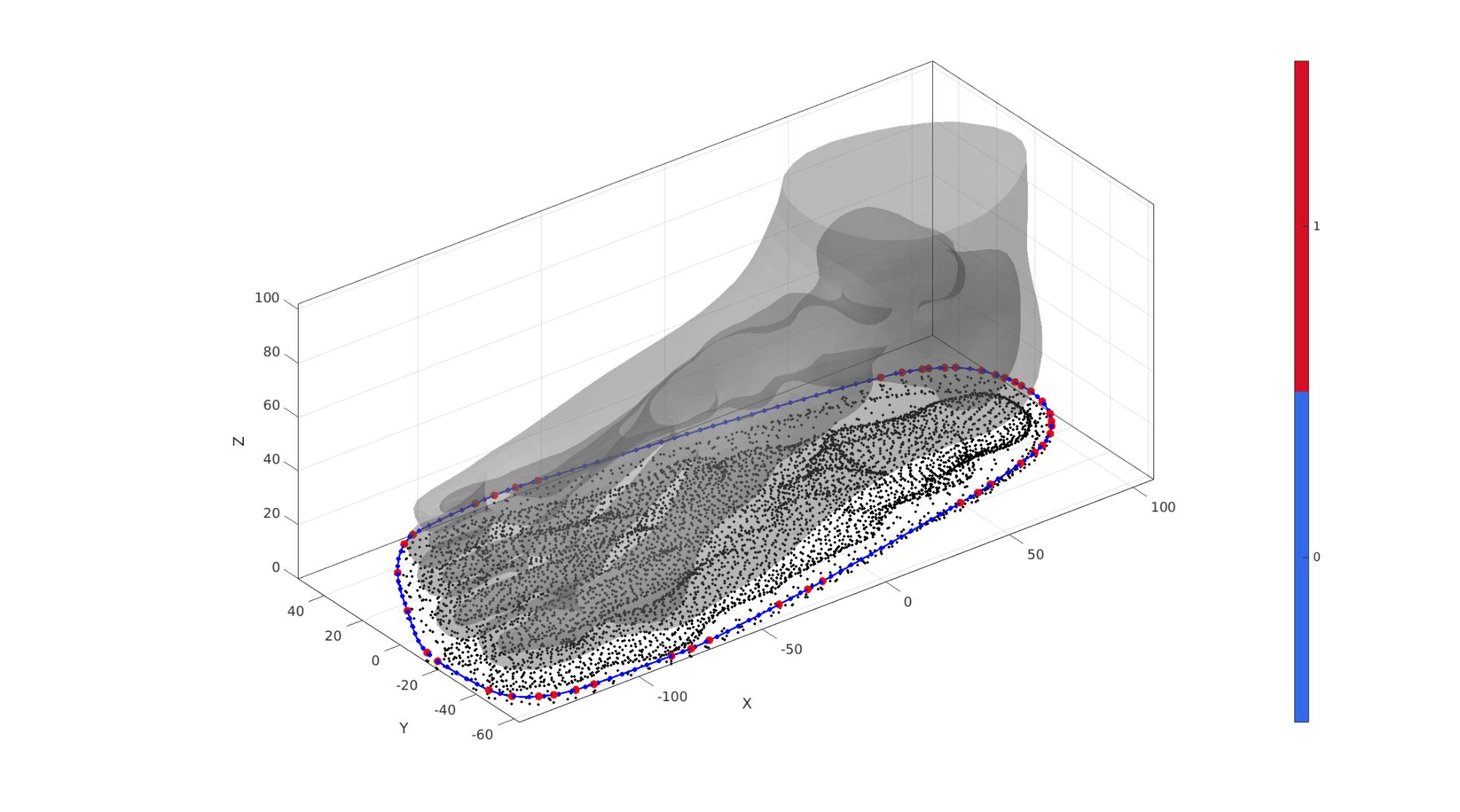
Create sole boundary curve in 2D
% Offset surface outward to thicken so sole is enlarged outward [~,~,Nv]=patchNormal(FT,VT); VT_sole=VT+soleOffsetOutward.*Nv; P=VT_sole(:,[1 2]); %Point set flattened to 2D DT=delaunayTriangulation(P); %Delaunay triangulation of 2D set VD=DT.Points; %Delaunay point set VD(:,3)=min(VT(:,3)); %Set z-coord to minimum for now indChull=DT.convexHull; %Ordered point list for conhex hull indChull=indChull(1:end-1); %Trim away last (=start) point to avoid double V_chull=VD(indChull,:); %Vertices for convex hull D=max(pathLength(V_chull)); %Get length of sole curve for resampling numResample=ceil(D./pointSpacingSole); V_sole_curve=evenlySampleCurve(V_chull,numResample,'pchip',1);
Visualize
cFigure; hold on; gpatch(FT,VT,'kw','none',0.25); plotV(P,'k.','MarkerSize',markerSize2); plotV(V_chull,'r.','MarkerSize',markerSize1*2); plotV(V_sole_curve,'b.-','MarkerSize',markerSize1,'LineWidth',lineWidth); camlight('headlight'); axisGeom(gca,fontSize); colormap gjet; icolorbar; drawnow;
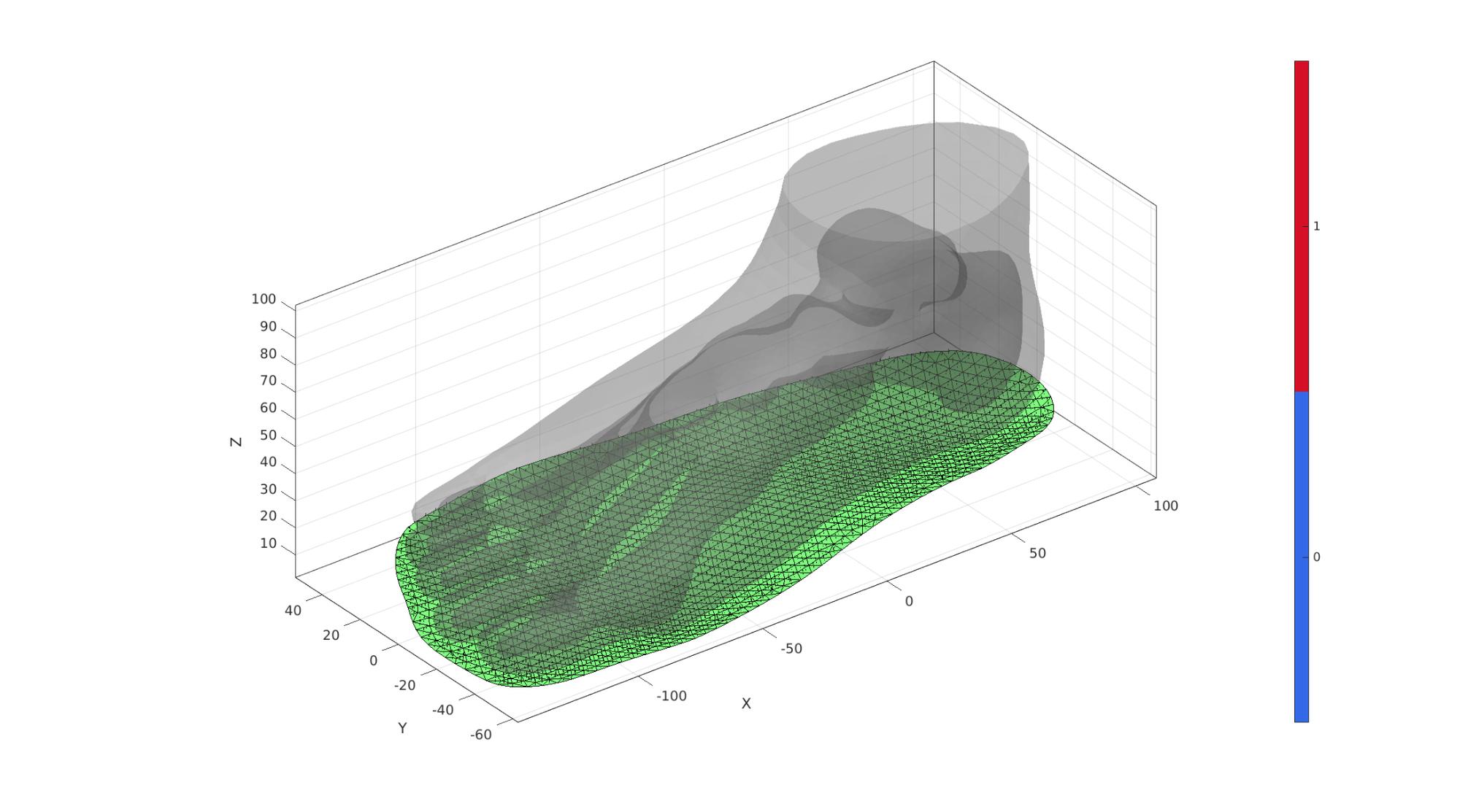
Build sole top surface
[F_sole_top,V_sole_top]=regionTriMesh2D({V_sole_curve(:,[1 2])},pointSpacingSole,0,0);
V_sole_top(:,3)=mean(V_sole_curve(:,3));
Eb=patchBoundary(F_sole_top);
indBoundary=unique(Eb(:));
%Get z-coordinate
[~,indMin]=minDist(V_sole_top,VT);
V_sole_top(:,3)=VT(indMin,3);
%Free smoothing of boundary
clear cParSmooth;
cParSmooth.n=numSmoothIterations_sole_Z;
cParSmooth.Method='LAP';
[p]=patchSmooth(Eb,V_sole_top,[],cParSmooth);
V_sole_top(indBoundary,3)=p(indBoundary,3);
% Constrained general smoothing
cParSmooth.n=numSmoothIterations_sole_Z;
cParSmooth.Method='LAP';
cParSmooth.RigidConstraints=indBoundary;
[V_sole_top(:,3)]=patchSmooth(F_sole_top,V_sole_top(:,3),[],cParSmooth);
%Shift a small amount to there is no initial contact
[~,indMin]=minDist(V_sole_top,VT);
dz=VT(indMin,3)-V_sole_top(:,3);
V_sole_top(:,3)=V_sole_top(:,3)-abs(min(dz));
V_sole_top(:,3)=V_sole_top(:,3)-initialSoleSpacing;
Visualize
cFigure; hold on; gpatch(FT,VT,'kw','none',0.25); gpatch(F_sole_top,V_sole_top,'gw','k',1); patchNormPlot(F_sole_top,V_sole_top); camlight('headlight'); axisGeom(gca,fontSize); colormap gjet; icolorbar; drawnow;
indBone=unique(Fb_foot(Cb_foot==1,:)); %Node numbers foot
D=minDist(V_sole_top,V_foot(indBone,:));
D=D-min(D(:));
D=D./max(D(:));
D=1-D;
spatVarThickness=(D*(soleMaxThickness-soleMinThickness))+soleMinThickness;
cFigure; hold on; gpatch(FT,VT,'kw','none',0.25); gpatch(F_sole_top,V_sole_top,spatVarThickness,'k',1); camlight('headlight'); axisGeom(gca,fontSize); colormap gjet; colorbar; drawnow;
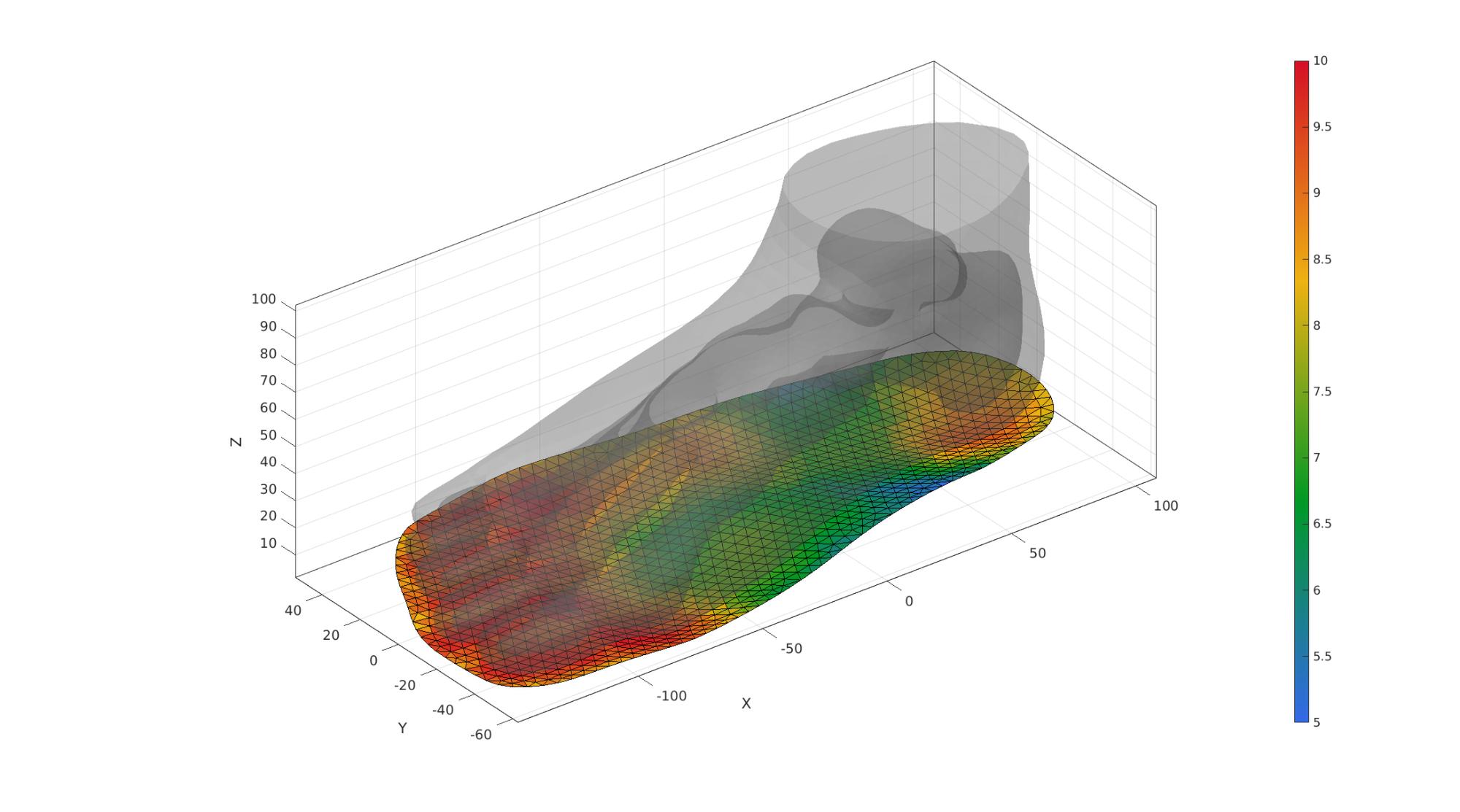
Create bottom surface of sole
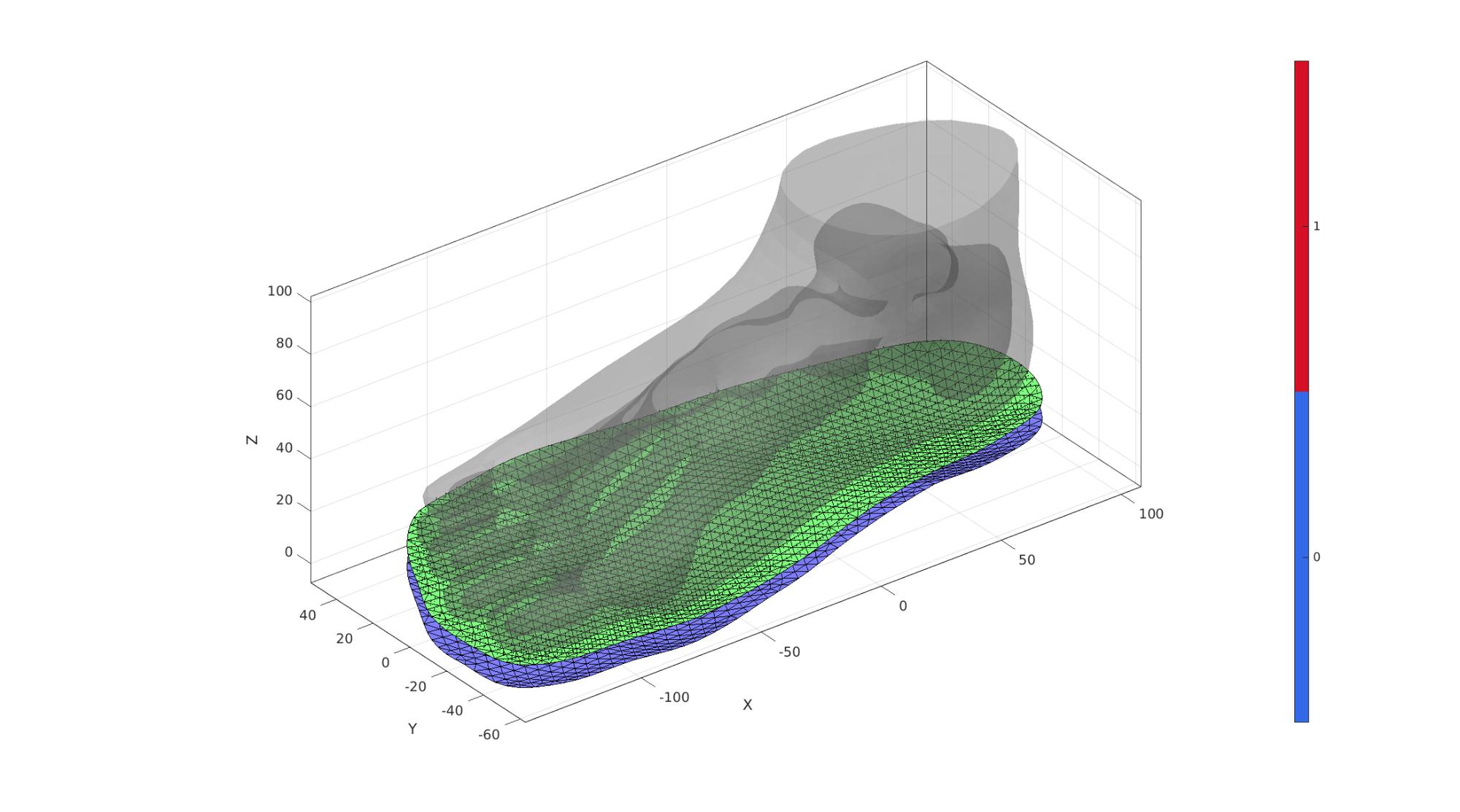
dirSet=[0 0 -1]; numElementsSoleThickness=ceil(soleMaxThickness./pointSpacingSole); [E_sole,V_sole,F_sole_top,F_sole_bottom]=patchThick(fliplr(F_sole_top),V_sole_top,dirSet,spatVarThickness,numElementsSoleThickness); F_sole_top=fliplr(F_sole_top); % Use element2patch to get patch data F_E_sole=element2patch(E_sole,[],'penta6'); % indBoundaryFaces=tesBoundary(F_E_sole,V_sole); % Fb_sole=F_E_sole(indBoundaryFaces,:);
cFigure; hold on; gpatch(FT,VT,'kw','none',0.25); gpatch(F_sole_top,V_sole,'gw','k',1); gpatch(F_sole_bottom,V_sole,'bw','k',1); patchNormPlot(F_sole_top,V_sole); camlight('headlight'); axisGeom(gca,fontSize); colormap gjet; icolorbar; drawnow;
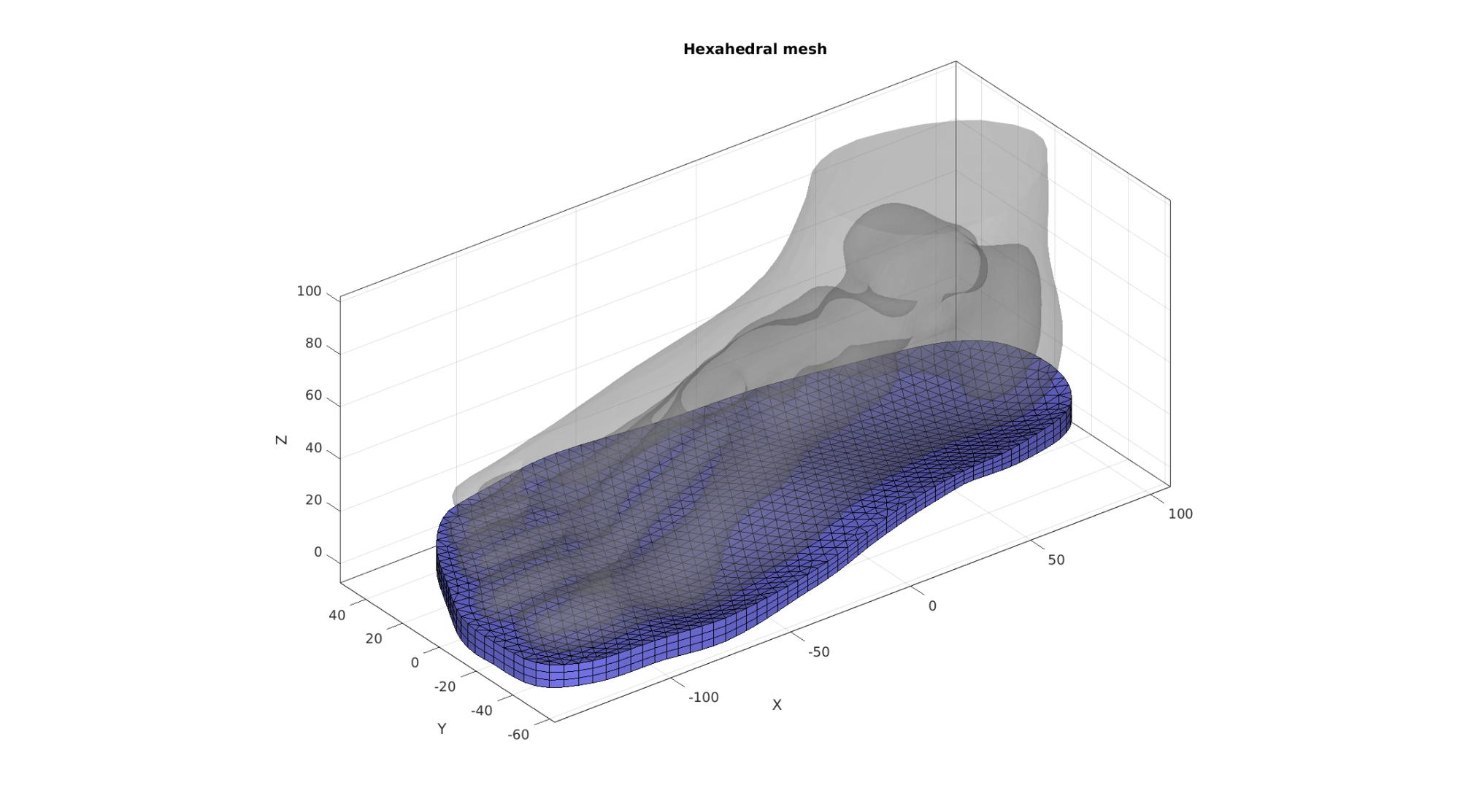
Visualize
cFigure; hold on; title('Hexahedral mesh'); gpatch(FT,VT,'kw','none',0.25); gpatch(F_E_sole,V_sole,'bw','k',1); % patchNormPlot(Fb_sole,V_sole); axisGeom; camlight headlight; drawnow;
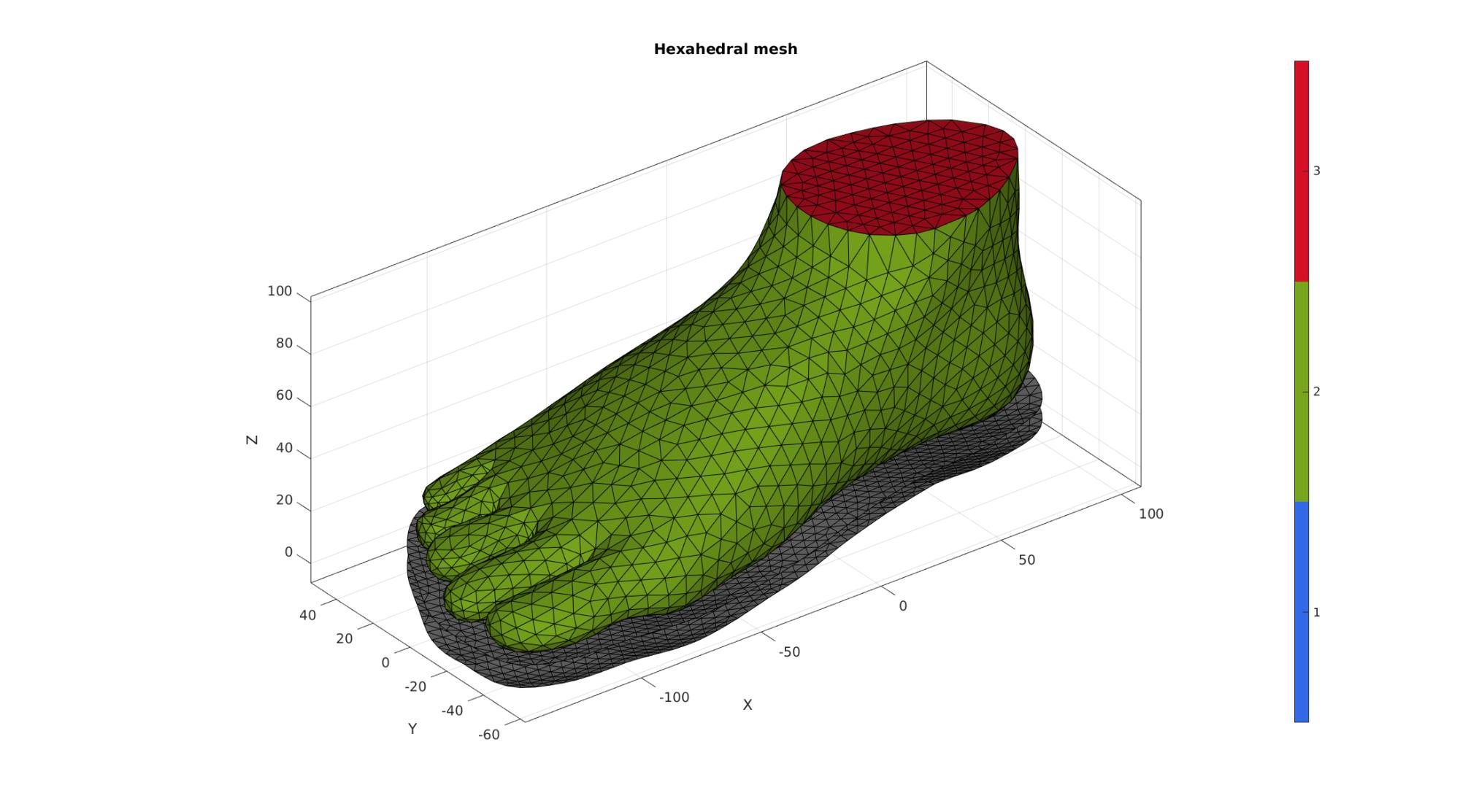
Joining node sets
V=[V_foot;V_sole;]; %Combined node sets E_sole=E_sole+size(V_foot,1); %Fixed element indices F_sole_top=F_sole_top+size(V_foot,1); %Fixed element indices F_sole_bottom=F_sole_bottom+size(V_foot,1); %Fixed indices F_E_sole=element2patch(E_sole,[],'penta6');
Visualize
cFigure; hold on; title('Hexahedral mesh'); gpatch(Fb_foot,V,Cb_foot,'k',1); gpatch(F_sole_top,V,'kw','k',1); gpatch(F_sole_bottom,V,'kw','k',1); % patchNormPlot(FEs,V); colormap gjet; icolorbar; axisGeom; camlight headlight; drawnow;
Define contact surfaces
% The rigid primary surface of the sphere F_contact_primary=F_sole_top; % The deformable secondary surface of the slab F_contact_secondary=fliplr(Fb_foot(Cb_foot==2,:));
Visualize contact surfaces
cFigure; hold on; title('Contact sets and normal directions','FontSize',fontSize); gpatch(Fb_foot,V,'kw','none',faceAlpha2); % gpatch(Fb_sole,V,'kw','none',faceAlpha2); hl(1)=gpatch(F_contact_primary,V,'gw','k',1); patchNormPlot(F_contact_primary,V); hl(2)=gpatch(F_contact_secondary,V,'bw','k',1); patchNormPlot(F_contact_secondary,V); legend(hl,{'Primary','Secondary'}); axisGeom(gca,fontSize); camlight headlight; drawnow;
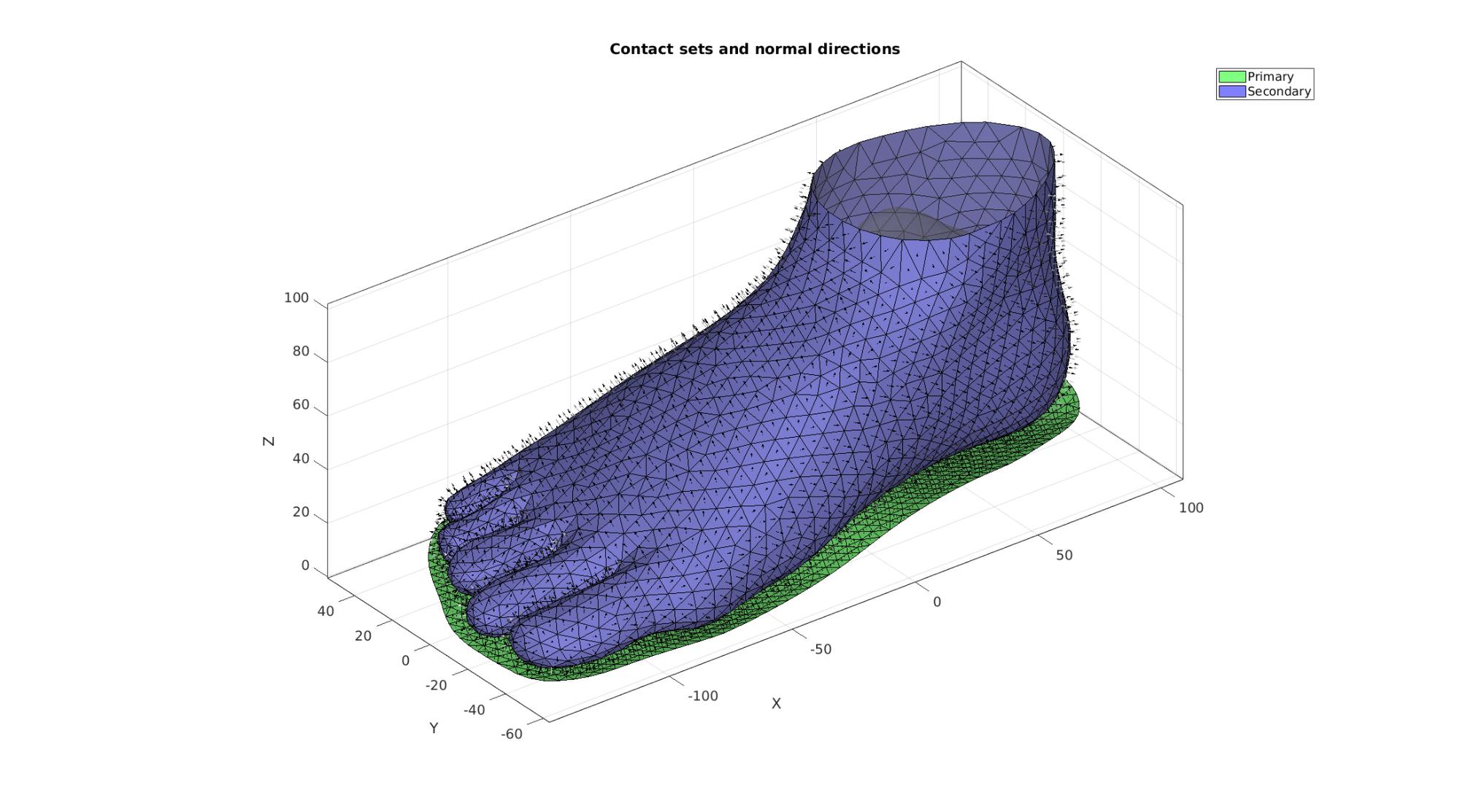
Define boundary conditions
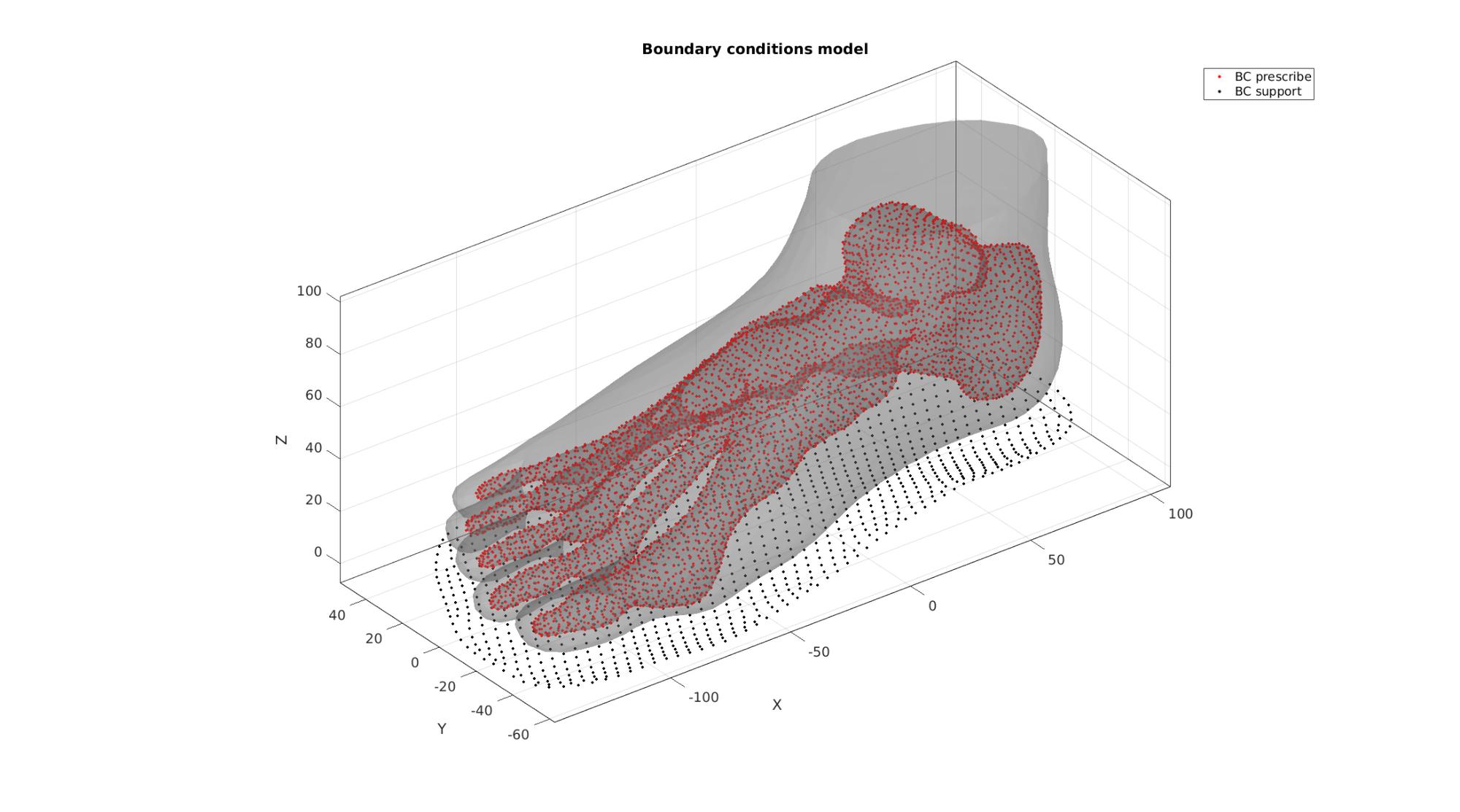
%Supported nodes bcSupportList=unique(F_sole_bottom); %Prescribed displacement nodes bcPrescribeList=unique(Fb_foot(Cb_foot==1,:));
Visualize BC's
hf=cFigure; hold on; title('Boundary conditions model','FontSize',fontSize); gpatch(Fb_foot,V,'kw','none',faceAlpha2); hl2(1)=plotV(V(bcPrescribeList,:),'r.','MarkerSize',markerSize2); hl2(2)=plotV(V(bcSupportList,:),'k.','MarkerSize',markerSize2); legend(hl2,{'BC prescribe','BC support'}); axisGeom(gca,fontSize); camlight headlight; drawnow;
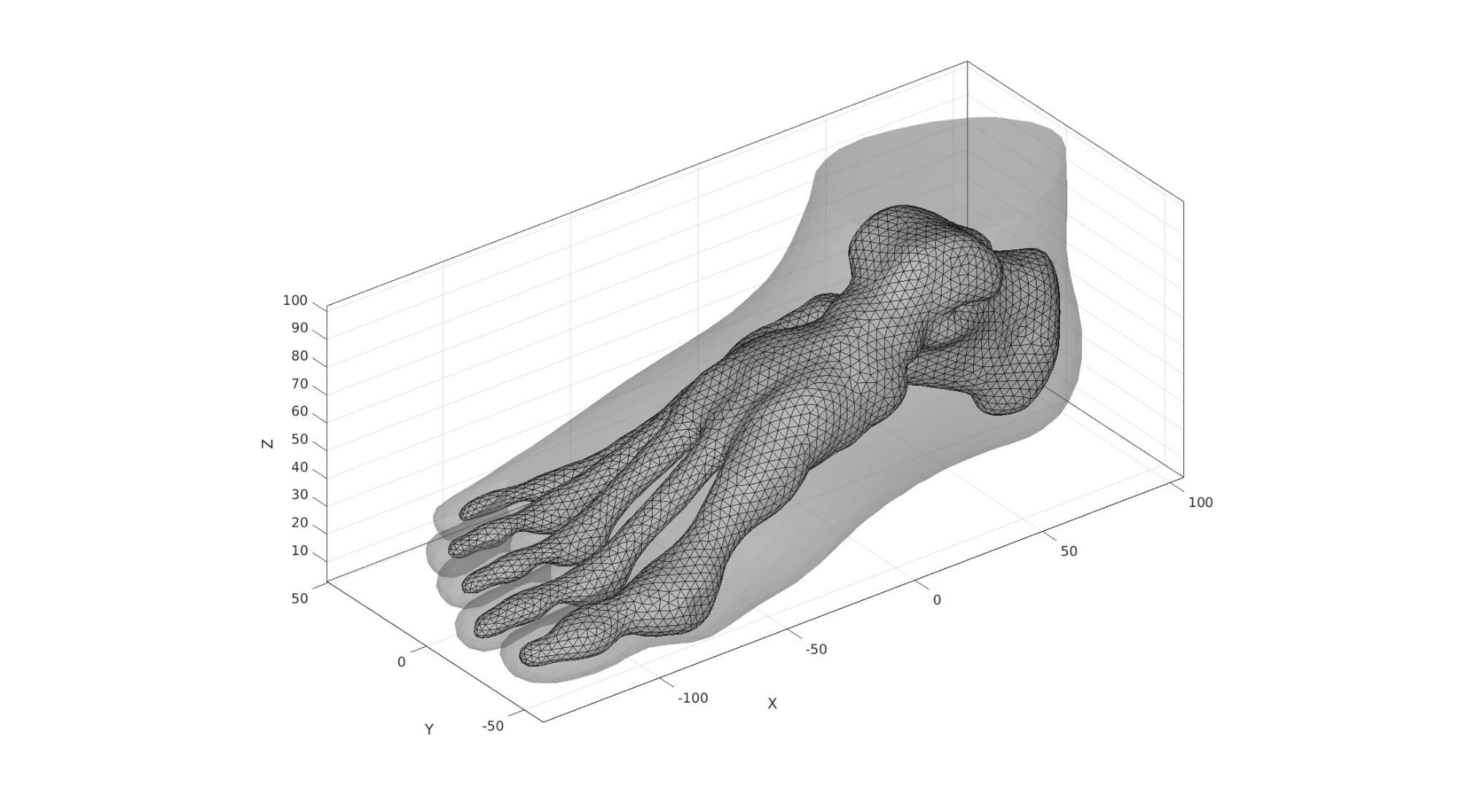
Visualize rigid body bone surface elements
F_bone=Fb_foot(Cb_foot==1,:); center_of_mass_bone=mean(V(bcPrescribeList,:),1); cFigure; hold on; % title('Boundary conditions model','FontSize',fontSize); gpatch(Fb_foot,V,'kw','none',faceAlpha2); gpatch(F_bone,V,'w','k',1); axisGeom(gca,fontSize); camlight headlight; drawnow;
Set-up materials
switch testCase case 0 %Spatially varying based on bone distance VE=patchCentre(E_sole,V); %Element centres V_vulnerable=VT(bcPrescribeList,:); W=minDist(VE,V_vulnerable); %Distance to bone points W=W-min(W(:)); W=W./max(W(:)); W=W.*(E_youngs_max-E_youngs_min); W=W+E_youngs_min; E_youngs_desired=W; case 1 % E_youngs_desired=E_youngs_max*ones(size(E_sole,1),1); case 2 E_youngs_desired=(E_youngs_max+E_youngs_min)/2*ones(size(E_sole,1),1); case 3 E_youngs_desired=E_youngs_min*ones(size(E_sole,1),1); end %Snap to nearest available materials [~,elementMaterialID]=minDist(E_youngs_desired(:),E_youngs_range(:));
Visualize material levels
%Use element2patch to get patch and color data [F_E_sole,C_F_E_sole]=element2patch(E_sole,elementMaterialID,'penta6'); hf=cFigure; hold on; title('Material levels','FontSize',fontSize); gpatch(Fb_foot(Cb_foot==1,:),V,'kw','none',1); gpatch(Fb_foot(Cb_foot~=1,:),V,'w','none',0.5); for q=1:1:numel(F_E_sole) gpatch(F_E_sole{q},V,C_F_E_sole{q},'k',1); end axisGeom(gca,fontSize); camlight headlight; colormap parula; icolorbar; %caxis([0 maxLevelColorbar_SED]); drawnow;
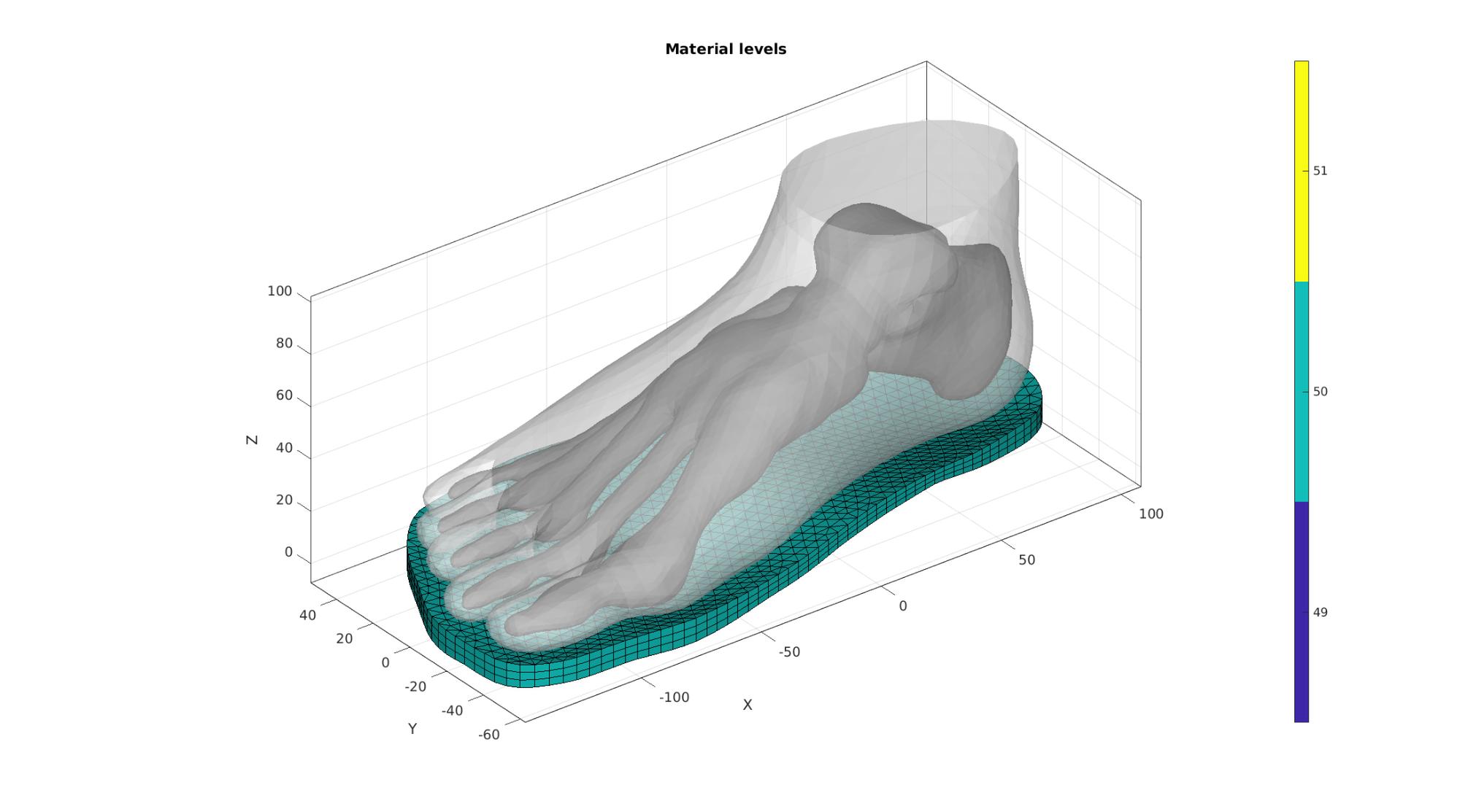
Convert list of materials to element id
%Sorting elements according to material label [elementMaterialID,indSortElements]=sort(elementMaterialID); E_sole=E_sole(indSortElements,:); %Removing unused materials [indUsed,ind1,ind2]=unique(elementMaterialID(:)); elementMaterialID=ind2; E_youngs_set=E_youngs_range(indUsed); numMaterials=numel(indUsed); E_youngs_elem=E_youngs_set(elementMaterialID);
Defining the FEBio input structure
See also febioStructTemplate and febioStruct2xml and the FEBio user manual.
%Get a template with default settings [febio_spec]=febioStructTemplate; %febio_spec version febio_spec.ATTR.version='4.0'; %Module section febio_spec.Module.ATTR.type='solid'; %Control section febio_spec.Control.analysis='STATIC'; febio_spec.Control.time_steps=numTimeSteps; febio_spec.Control.step_size=1/numTimeSteps; febio_spec.Control.solver.max_refs=max_refs; febio_spec.Control.solver.qn_method.max_ups=max_ups; febio_spec.Control.solver.symmetric_stiffness=symmetric_stiffness; febio_spec.Control.time_stepper.dtmin=dtmin; febio_spec.Control.time_stepper.dtmax=dtmax; febio_spec.Control.time_stepper.max_retries=max_retries; febio_spec.Control.time_stepper.opt_iter=opt_iter; %Material section materialName1='Material1'; febio_spec.Material.material{1}.ATTR.name=materialName1; febio_spec.Material.material{1}.ATTR.type='Ogden'; febio_spec.Material.material{1}.ATTR.id=1; febio_spec.Material.material{1}.c1=c1_1; febio_spec.Material.material{1}.m1=m1_1; febio_spec.Material.material{1}.k=k_1; materialName2='Material2'; febio_spec.Material.material{2}.ATTR.name=materialName2; febio_spec.Material.material{2}.ATTR.type='rigid body'; febio_spec.Material.material{2}.ATTR.id=2; febio_spec.Material.material{2}.density=1; febio_spec.Material.material{2}.center_of_mass=center_of_mass_bone; materialName3='Material3'; dataMapName='MaterialParameterMap'; febio_spec.Material.material{3}.ATTR.name=materialName3; febio_spec.Material.material{3}.ATTR.type='neo-Hookean'; febio_spec.Material.material{3}.ATTR.id=3; febio_spec.Material.material{3}.E.ATTR.type='map'; %Calls for mapping of parameter febio_spec.Material.material{3}.E=dataMapName; %Calls for mapping of parameter febio_spec.Material.material{3}.v=nu; % Mesh section % -> Nodes febio_spec.Mesh.Nodes{1}.ATTR.name='Object1'; %The node set name febio_spec.Mesh.Nodes{1}.node.ATTR.id=(1:size(V,1))'; %The node id's febio_spec.Mesh.Nodes{1}.node.VAL=V; %The nodel coordinates % -> Elements partName1='Part1'; febio_spec.Mesh.Elements{1}.ATTR.name=partName1; %Name of this part febio_spec.Mesh.Elements{1}.ATTR.type='tet4'; %Element type febio_spec.Mesh.Elements{1}.elem.ATTR.id=(1:1:size(E_foot,1))'; %Element id's febio_spec.Mesh.Elements{1}.elem.VAL=E_foot; %The element matrix partName2='Part2'; febio_spec.Mesh.Elements{2}.ATTR.name=partName2; %Name of this part febio_spec.Mesh.Elements{2}.ATTR.type='tri3'; %Element type febio_spec.Mesh.Elements{2}.elem.ATTR.id=size(E_foot,1)+(1:1:size(F_bone,1))'; %Element id's febio_spec.Mesh.Elements{2}.elem.VAL=F_bone; %The element matrix partName3='Part3'; febio_spec.Mesh.Elements{3}.ATTR.name=partName3; %Name of this part febio_spec.Mesh.Elements{3}.ATTR.type='penta6'; %Element type febio_spec.Mesh.Elements{3}.elem.ATTR.id=size(E_foot,1)+size(F_bone,1)+(1:1:size(E_sole,1))'; %Element id's febio_spec.Mesh.Elements{3}.elem.VAL=E_sole; %The element matrix % -> NodeSets nodeSetName1='bcSupportList'; nodeSetName2='bcPrescribeList'; febio_spec.Mesh.NodeSet{1}.ATTR.name=nodeSetName1; febio_spec.Mesh.NodeSet{1}.VAL=mrow(bcSupportList); febio_spec.Mesh.NodeSet{2}.ATTR.name=nodeSetName2; febio_spec.Mesh.NodeSet{2}.VAL=mrow(bcPrescribeList); % -> Surfaces surfaceName1='contactSurface1'; febio_spec.Mesh.Surface{1}.ATTR.name=surfaceName1; febio_spec.Mesh.Surface{1}.tri3.ATTR.id=(1:1:size(F_contact_secondary,1))'; febio_spec.Mesh.Surface{1}.tri3.VAL=F_contact_secondary; surfaceName2='contactSurface2'; febio_spec.Mesh.Surface{2}.ATTR.name=surfaceName2; febio_spec.Mesh.Surface{2}.tri3.ATTR.id=(1:1:size(F_contact_primary,1))'; febio_spec.Mesh.Surface{2}.tri3.VAL=F_contact_primary; % -> Surface pairs contactPairName='Contact1'; febio_spec.Mesh.SurfacePair{1}.ATTR.name=contactPairName; febio_spec.Mesh.SurfacePair{1}.primary=surfaceName2; febio_spec.Mesh.SurfacePair{1}.secondary=surfaceName1; %MeshData secion %-> Element data febio_spec.MeshData.ElementData.ATTR.name=dataMapName; febio_spec.MeshData.ElementData.ATTR.elem_set=partName3; febio_spec.MeshData.ElementData.elem.ATTR.lid=(1:1:size(E_sole,1))'; febio_spec.MeshData.ElementData.elem.VAL=E_youngs_elem(:); %MeshDomains section febio_spec.MeshDomains.SolidDomain{1}.ATTR.name=partName1; febio_spec.MeshDomains.SolidDomain{1}.ATTR.mat=materialName1; febio_spec.MeshDomains.ShellDomain.ATTR.name=partName2; febio_spec.MeshDomains.ShellDomain.ATTR.mat=materialName2; febio_spec.MeshDomains.SolidDomain{2}.ATTR.name=partName3; febio_spec.MeshDomains.SolidDomain{2}.ATTR.mat=materialName3; %Boundary condition section % -> Fix boundary conditions febio_spec.Boundary.bc{1}.ATTR.name='zero_displacement_xyz'; febio_spec.Boundary.bc{1}.ATTR.type='zero displacement'; febio_spec.Boundary.bc{1}.ATTR.node_set=nodeSetName1; febio_spec.Boundary.bc{1}.x_dof=1; febio_spec.Boundary.bc{1}.y_dof=1; febio_spec.Boundary.bc{1}.z_dof=1; %Rigid section % ->Rigid body fix boundary conditions febio_spec.Rigid.rigid_bc{1}.ATTR.name='RigidFix'; febio_spec.Rigid.rigid_bc{1}.ATTR.type='rigid_fixed'; febio_spec.Rigid.rigid_bc{1}.rb=2; febio_spec.Rigid.rigid_bc{1}.Rx_dof=1; febio_spec.Rigid.rigid_bc{1}.Ry_dof=1; febio_spec.Rigid.rigid_bc{1}.Ru_dof=1; febio_spec.Rigid.rigid_bc{1}.Rv_dof=1; febio_spec.Rigid.rigid_bc{1}.Rw_dof=1; % ->Rigid body prescribe boundary conditions febio_spec.Rigid.rigid_bc{2}.ATTR.name='RigidPrescribe'; febio_spec.Rigid.rigid_bc{2}.ATTR.type='rigid_displacement'; febio_spec.Rigid.rigid_bc{2}.rb=2; febio_spec.Rigid.rigid_bc{2}.dof='z'; febio_spec.Rigid.rigid_bc{2}.value.ATTR.lc=1; febio_spec.Rigid.rigid_bc{2}.value.VAL=displacementMagnitude; febio_spec.Rigid.rigid_bc{2}.relative=0; %Contact section febio_spec.Contact.contact{1}.ATTR.type='sliding-elastic'; febio_spec.Contact.contact{1}.ATTR.surface_pair=contactPairName; febio_spec.Contact.contact{1}.two_pass=0; febio_spec.Contact.contact{1}.laugon=laugon; febio_spec.Contact.contact{1}.tolerance=0.2; febio_spec.Contact.contact{1}.gaptol=0; febio_spec.Contact.contact{1}.minaug=minaug; febio_spec.Contact.contact{1}.maxaug=maxaug; febio_spec.Contact.contact{1}.search_tol=0.01; febio_spec.Contact.contact{1}.search_radius=0.1*sqrt(sum((max(V,[],1)-min(V,[],1)).^2,2)); febio_spec.Contact.contact{1}.symmetric_stiffness=0; febio_spec.Contact.contact{1}.auto_penalty=1; febio_spec.Contact.contact{1}.penalty=contactPenalty; febio_spec.Contact.contact{1}.fric_coeff=fric_coeff; %LoadData section % -> load_controller febio_spec.LoadData.load_controller{1}.ATTR.name='LC_1'; febio_spec.LoadData.load_controller{1}.ATTR.id=1; febio_spec.LoadData.load_controller{1}.ATTR.type='loadcurve'; febio_spec.LoadData.load_controller{1}.interpolate='LINEAR'; %febio_spec.LoadData.load_controller{1}.extend='CONSTANT'; febio_spec.LoadData.load_controller{1}.points.pt.VAL=[0 0; 1 1]; %Output section % -> log file febio_spec.Output.logfile.ATTR.file=febioLogFileName; febio_spec.Output.logfile.node_data{1}.ATTR.file=febioLogFileName_disp; febio_spec.Output.logfile.node_data{1}.ATTR.data='ux;uy;uz'; febio_spec.Output.logfile.node_data{1}.ATTR.delim=','; febio_spec.Output.logfile.rigid_body_data{1}.ATTR.file=febioLogFileName_force; febio_spec.Output.logfile.rigid_body_data{1}.ATTR.data='Fx;Fy;Fz'; febio_spec.Output.logfile.rigid_body_data{1}.ATTR.delim=','; febio_spec.Output.logfile.rigid_body_data{1}.VAL=2; febio_spec.Output.logfile.element_data{1}.ATTR.file=febioLogFileName_sed; febio_spec.Output.logfile.element_data{1}.ATTR.data='sed'; febio_spec.Output.logfile.element_data{1}.ATTR.delim=','; % Plotfile section febio_spec.Output.plotfile.compression=0;
Quick viewing of the FEBio input file structure
The febView function can be used to view the xml structure in a MATLAB figure window.
febView(febio_spec); %Viewing the febio file
Running the FEBio analysis
To run the analysis defined by the created FEBio input file the runMonitorFEBio function is used. The input for this function is a structure defining job settings e.g. the FEBio input file name. The optional output runFlag informs the user if the analysis was run succesfully.
febioAnalysis.run_filename=febioFebFileName; %The input file name febioAnalysis.run_logname=febioLogFileName; %The name for the log file febioAnalysis.disp_on=1; %Display information on the command window febioAnalysis.runMode=runMode;
if optimizeForceOption==0
Exporting the FEBio input file
Exporting the febio_spec structure to an FEBio input file is done using the febioStruct2xml function.
febioStruct2xml(febio_spec,febioFebFileName); %Exporting to file and domNode
Run febio
[runFlag]=runMonitorFEBio(febioAnalysis);%START FEBio NOW!!!!!!!! if runFlag~=1 error('FEBio error'); end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
--------> RUNNING/MONITORING FEBIO JOB <-------- 27-Apr-2023 16:18:25
FEBio path: /home/kevin/FEBioStudio2/bin/febio4
# Attempt removal of existing log files 27-Apr-2023 16:18:25
* Removal succesful 27-Apr-2023 16:18:25
# Attempt removal of existing .xplt files 27-Apr-2023 16:18:26
* Removal succesful 27-Apr-2023 16:18:26
# Starting FEBio... 27-Apr-2023 16:18:26
Max. total analysis time is: Inf s
* Waiting for log file creation 27-Apr-2023 16:18:26
Max. wait time: 30 s
* Log file found. 27-Apr-2023 16:18:26
# Parsing log file... 27-Apr-2023 16:18:26
number of iterations : 1 27-Apr-2023 16:18:27
number of reformations : 1 27-Apr-2023 16:18:27
------- converged at time : 0.1 27-Apr-2023 16:18:27
number of iterations : 4 27-Apr-2023 16:18:31
number of reformations : 4 27-Apr-2023 16:18:31
------- converged at time : 0.2 27-Apr-2023 16:18:31
number of iterations : 5 27-Apr-2023 16:18:35
number of reformations : 5 27-Apr-2023 16:18:35
------- converged at time : 0.3 27-Apr-2023 16:18:35
number of iterations : 5 27-Apr-2023 16:18:39
number of reformations : 5 27-Apr-2023 16:18:39
------- converged at time : 0.4 27-Apr-2023 16:18:39
number of iterations : 16 27-Apr-2023 16:18:50
number of reformations : 16 27-Apr-2023 16:18:50
------- converged at time : 0.5 27-Apr-2023 16:18:50
number of iterations : 6 27-Apr-2023 16:18:55
number of reformations : 6 27-Apr-2023 16:18:55
------- converged at time : 0.579266 27-Apr-2023 16:18:55
number of iterations : 9 27-Apr-2023 16:19:02
number of reformations : 9 27-Apr-2023 16:19:02
------- converged at time : 0.662679 27-Apr-2023 16:19:02
number of iterations : 6 27-Apr-2023 16:19:07
number of reformations : 6 27-Apr-2023 16:19:07
------- converged at time : 0.74699 27-Apr-2023 16:19:07
number of iterations : 8 27-Apr-2023 16:19:14
number of reformations : 8 27-Apr-2023 16:19:14
------- converged at time : 0.834438 27-Apr-2023 16:19:14
number of iterations : 15 27-Apr-2023 16:19:27
number of reformations : 15 27-Apr-2023 16:19:27
------- converged at time : 0.923368 27-Apr-2023 16:19:27
number of iterations : 12 27-Apr-2023 16:19:37
number of reformations : 12 27-Apr-2023 16:19:37
------- converged at time : 0.996162 27-Apr-2023 16:19:37
number of iterations : 7 27-Apr-2023 16:19:43
number of reformations : 7 27-Apr-2023 16:19:43
------- converged at time : 1 27-Apr-2023 16:19:43
Elapsed time : 0:01:17 27-Apr-2023 16:19:44
N O R M A L T E R M I N A T I O N
# Done 27-Apr-2023 16:19:44
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
elseif optimizeForceOption==1 while 1
Set displacement magnitude in input file structure
febio_spec.Rigid.rigid_constraint{2}.value.VAL=displacementMagnitude;
Export input file
Exporting the febio_spec structure to an FEBio input file is done using the febioStruct2xml function.
febioStruct2xml(febio_spec,febioFebFileName); %Exporting to file and domNode
Run febio
[runFlag]=runMonitorFEBio(febioAnalysis);%START FEBio NOW!!!!!!!! if runFlag~=1 error('FEBio error'); end
Importing rigid body reaction forces from a log file
dataStructForce=importFEBio_logfile(fullfile(savePath,febioLogFileName_force),0,1);
F_reaction=squeeze(dataStructForce.data(1,:,:))';
timeVec=dataStructForce.time;
Fz_final=F_reaction(end,3);
forceDifference=abs(Fz_final-forceBody);
Use inter/extra-polation to guess displacement
u=timeVec.*displacementMagnitude;
f=F_reaction(:,3);
displacementProposed=interp1(f,u,forceBody,'linear','extrap');
cFigure; hold on; hp(1)=plot(f,u,'r-','LineWidth',lineWidth); hp(2)=plot(forceBody,displacementProposed,'b.','MarkerSize',25); hl=legend(hp,{'displacement - force FEA','Proposed displacement'},'FontSize',fontSize); axis square; axis tight; grid on; box on; drawnow;
displacementDifference=displacementProposed-displacementMagnitude;
if displacementDifference<-maxProposedDisplacementDifference
displacementMagnitude=displacementMagnitude-maxProposedDisplacementDifference;
else
displacementMagnitude=displacementProposed;
end
disp(['Force is: ',num2str(Fz_final),' N. Setting displacement to: ',num2str(displacementMagnitude)]); if forceDifference<forceDifferenceTolerance break end
end end
Import rigid body reaction forces
dataStructForce=importFEBio_logfile(fullfile(savePath,febioLogFileName_force),0,1); F_reaction=squeeze(dataStructForce.data(1,:,:))'; timeVec=dataStructForce.time;
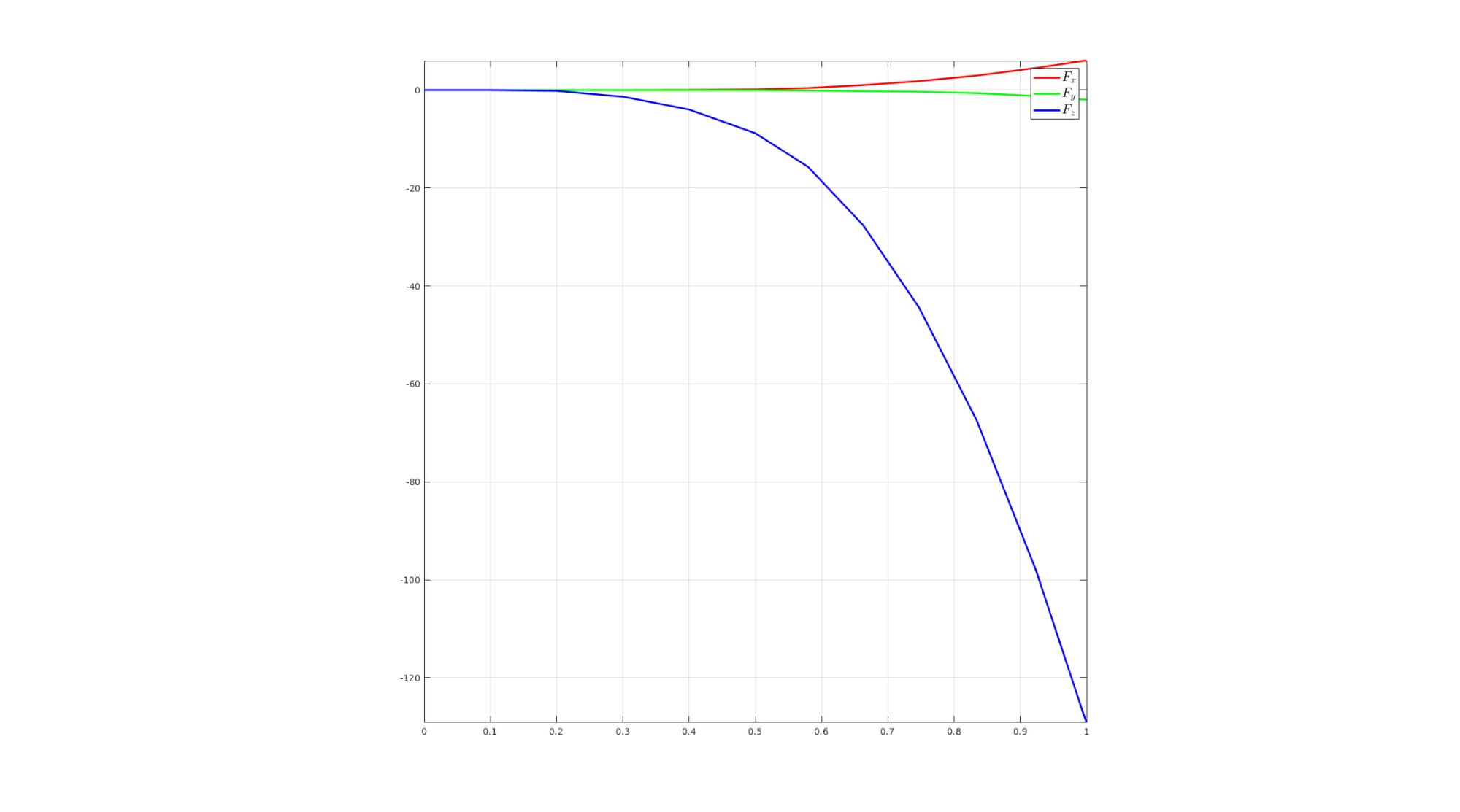
Visualize force
cFigure; hold on; hp(1)=plot(timeVec,F_reaction(:,1),'r-','LineWidth',lineWidth); hp(2)=plot(timeVec,F_reaction(:,2),'g-','LineWidth',lineWidth); hp(3)=plot(timeVec,F_reaction(:,3),'b-','LineWidth',lineWidth); hl=legend(hp,{'$F_x$','$F_y$','$F_z$'},'Interpreter','Latex','FontSize',fontSize); axis square; axis tight; grid on; box on; drawnow;
Importing nodal displacements from a log file
dataStructDisp=importFEBio_logfile(fullfile(savePath,febioLogFileName_disp),0,1); %Access data N_disp_mat=dataStructDisp.data; %Displacement timeVec=dataStructDisp.time; %Time %Create deformed coordinate set V_DEF=N_disp_mat+repmat(V,[1 1 size(N_disp_mat,3)]);
Importing element stress from a log file
dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_sed),0,1);
%Access data
E_sed_mat=dataStruct.data;
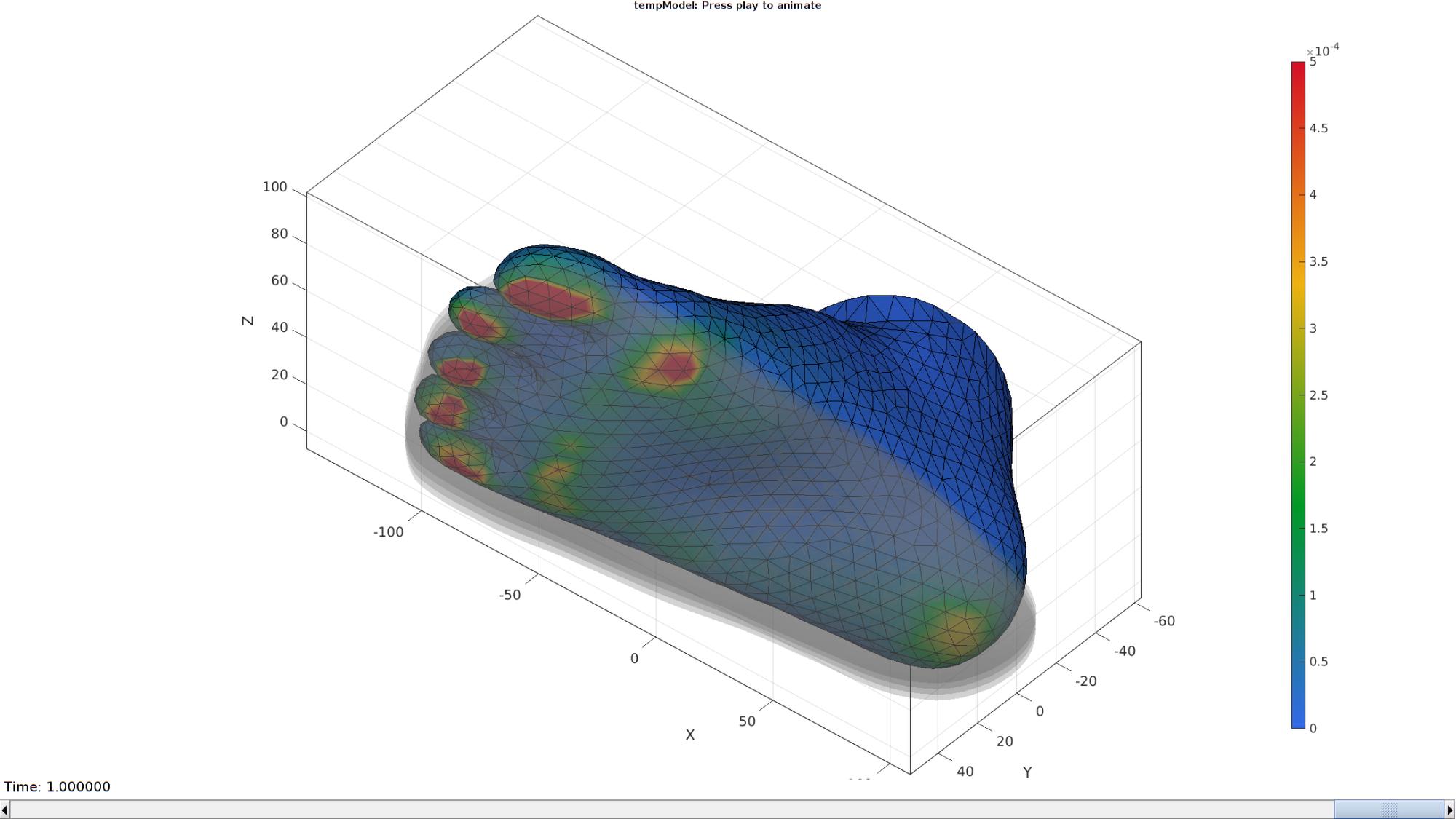
Plot animation
Plotting the simulated results using anim8 to visualize and animate deformations
[CV]=faceToVertexMeasure(E_foot,V,E_sed_mat(:,:,end)); % Create basic view and store graphics handle to initiate animation hf=cFigure; hold on; %Open figure gtitle([febioFebFileNamePart,': Press play to animate']); hp1=gpatch(Fb_foot,V_DEF(:,:,end),CV,'k',1); %Add graphics object to animate hp1.FaceColor='Interp'; hp2=gpatch(F_E_sole{1},V_DEF(:,:,end),'kw','none',0.25); %Add graphics object to animate axisGeom(gca,fontSize); colormap(gjet(250)); colorbar; % caxis([0 max(C_energy_foot)/100]); caxis([0 maxLevelColorbar_SED]); axis(axisLim(V_DEF)); %Set axis limits statically view([-40 -40]); camlight headlight; % Set up animation features animStruct.Time=timeVec; %The time vector for qt=1:1:size(V_DEF,3) %Loop over time increments [CV]=faceToVertexMeasure(E_foot,V,E_sed_mat(:,:,qt)); %Set entries in animation structure animStruct.Handles{qt}=[hp1 hp1 hp2]; %Handles of objects to animate animStruct.Props{qt}={'Vertices','CData','Vertices'}; %Properties of objects to animate animStruct.Set{qt}={V_DEF(:,:,qt),CV,V_DEF(:,:,qt)}; %Property values for to set in order to animate end anim8(hf,animStruct); %Initiate animation feature drawnow;

GIBBON www.gibboncode.org
Kevin Mattheus Moerman, [email protected]
GIBBON footer text
License: https://github.com/gibbonCode/GIBBON/blob/master/LICENSE
GIBBON: The Geometry and Image-based Bioengineering add-On. A toolbox for image segmentation, image-based modeling, meshing, and finite element analysis.
Copyright (C) 2006-2022 Kevin Mattheus Moerman and the GIBBON contributors
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.