DEMO_febio_0035_blob_shear_contact_hex8
Below is a demonstration for:
- Building geometry for a hemi-spherical blob with hexahedral elements which is being sheared by a rigid wall. This demo consists off:
- Defining the boundary conditions
- Coding the febio structure
- Running the model
- Importing and visualizing the displacement results
Contents
- Keywords
- Plot settings
- Control parameters
- Creating model geometry and mesh
- Creating rigid body shear surface
- Join model node sets
- Get local material axes
- Visualize contact surfaces
- Setup boundary conditions
- Defining the FEBio input structure
- Quick viewing of the FEBio input file structure
- Exporting the FEBio input file
- Running the FEBio analysis
- Import FEBio results
Keywords
- febio_spec version 4.0
- febio, FEBio
- indentation
- contact, sliding, friction
- rigid body constraints
- hexahedral elements, hex8, hex20
- quadrilaterial elements, quad4, quad8
- shell elements
- sphere
- static, solid
- hyperelastic, Ogden
- hyperelastic, Fung orthotropic
- displacement logfile
clear; close all; clc;
Plot settings
fontSize=15; faceAlpha1=0.8; faceAlpha2=0.3; markerSize=40; markerSize2=15; lineWidth=3;
Control parameters
% Path names defaultFolder = fileparts(fileparts(mfilename('fullpath'))); savePath=fullfile(defaultFolder,'data','temp'); % Defining file names febioFebFileNamePart='tempModel'; febioFebFileName=fullfile(savePath,[febioFebFileNamePart,'.feb']); %FEB file name febioLogFileName=fullfile(savePath,[febioFebFileNamePart,'.txt']); %FEBio log file name febioLogFileName_disp=[febioFebFileNamePart,'_disp_out.txt']; %Log file name for exporting displacement % Hemi-sphere parameters hemiSphereRadius=6; numElementsMantel=4; smoothEdge=1; solidElementType='hex8';%'hex20'; membraneThickness=0.1; % Ground plate parameters plateRadius=1.3*hemiSphereRadius; % Probe parameters probeWidth=3*hemiSphereRadius; filletProbe=hemiSphereRadius/2; %Fillet radius % Define probe displacement probeDisplacement=hemiSphereRadius*2; probeOverlapFactor=0.3; probeLength=hemiSphereRadius*2; % Material parameter set l=1e-3; lt=1000*l; m=1e-3; mt=1000*m; mu1=mt; mu2=mt; mu3=m; lambda11=lt; lambda22=lt; lambda12=lt; lambda33=l; lambda23=l; lambda13=l; [E1,E2,E3,G12,G23,G31,v12,v23,v31]=lameInvertHookeOrthotropic(mu1,mu2,mu3,lambda11,lambda22,lambda33,lambda12,lambda23,lambda13); materialPropertiesFung.E1=E1; materialPropertiesFung.E2=E2; materialPropertiesFung.E3=E3; materialPropertiesFung.G12=G12; materialPropertiesFung.G23=G23; materialPropertiesFung.G31=G31; materialPropertiesFung.v12=v12; materialPropertiesFung.v23=v23; materialPropertiesFung.v31=v31; materialPropertiesFung.c=10; materialPropertiesFung.k=1000*mean([G12 G23 G31]); %Ogden parameters materialPropertiesOgden.c1=1e-3; %Shear-modulus-like parameter materialPropertiesOgden.m1=2; %Material parameter setting degree of non-linearity materialPropertiesOgden.k=1000*materialPropertiesOgden.c1; %Bulk modulus % FEA control settings numTimeSteps=25; max_refs=25; %Max reforms max_ups=0; %Set to zero to use full-Newton iterations opt_iter=15; %Optimum number of iterations max_retries=25; %Maximum number of retires symmetric_stiffness=0; min_residual=1e-20; step_size=1/numTimeSteps; dtmin=(1/numTimeSteps)/100; %Minimum time step size dtmax=(1/numTimeSteps); %Maximum time step size runMode='internal';% 'internal' or 'external' %Contact parameters contactPenalty=0.01; laugon=0; minaug=1; maxaug=10; fric_coeff=0; max_traction=0;
Creating model geometry and mesh
%Control settings optionStruct.sphereRadius=hemiSphereRadius; optionStruct.coreRadius=optionStruct.sphereRadius/2; optionStruct.numElementsMantel=numElementsMantel; optionStruct.numElementsCore=optionStruct.numElementsMantel*2; optionStruct.outputStructType=2; optionStruct.makeHollow=0; optionStruct.cParSmooth.n=25; % %Creating sphere [meshStruct]=hexMeshHemiSphere(optionStruct); % Access model element and patch data Fb_blob=meshStruct.facesBoundary; Cb_blob=meshStruct.boundaryMarker; V_blob=meshStruct.nodes; E_blob=meshStruct.elements; F_blob=element2patch(E_blob);
pointSpacingBlob=max(patchEdgeLengths(Fb_blob,V_blob)); %Smoothen edges if smoothEdge==1 %Get rigid region ind=1:1:size(V_blob,1); %Indices for all nodes indRigid1=find(ismember(ind,Fb_blob(Cb_blob==2,:)) & ~ismember(ind,Fb_blob(Cb_blob==1,:))); %Indices for new bottom surface nodes indRigid2=find(ismember(ind,Fb_blob(Cb_blob==1,:)) & ~ismember(ind,Fb_blob(Cb_blob==2,:))); %Indices for new bottom surface nodes indRigid=[indRigid1(:); indRigid2(:);]; %Smoothing cPar.Method='HC'; cPar.n=250; cPar.RigidConstraints=indRigid; [Vb_blob]=patchSmooth(F_blob,V_blob,[],cPar); indSmooth=unique(F_blob(:)); V_blob(indSmooth,:)=Vb_blob(indSmooth,:); %Fix color data with new bottom surface Cb_blob=ones(size(Cb_blob)); Cb_blob(all(ismember(Fb_blob,indRigid1),2))=2; meshStruct.nodes=V_blob; end if strcmp(solidElementType,'hex20') [E_blob,V_blob,~,~,Fb_blob]=hex8_hex20(E_blob,V_blob,{},Fb_blob); [Fb_blob_plot]=element2patch(Fb_blob,[],'quad8'); meshStruct.elements=E_blob; meshStruct.nodes=V_blob; meshStruct.Fb=Fb_blob_plot; shellElementType='quad8'; else Fb_blob_plot=Fb_blob; shellElementType='quad4'; end
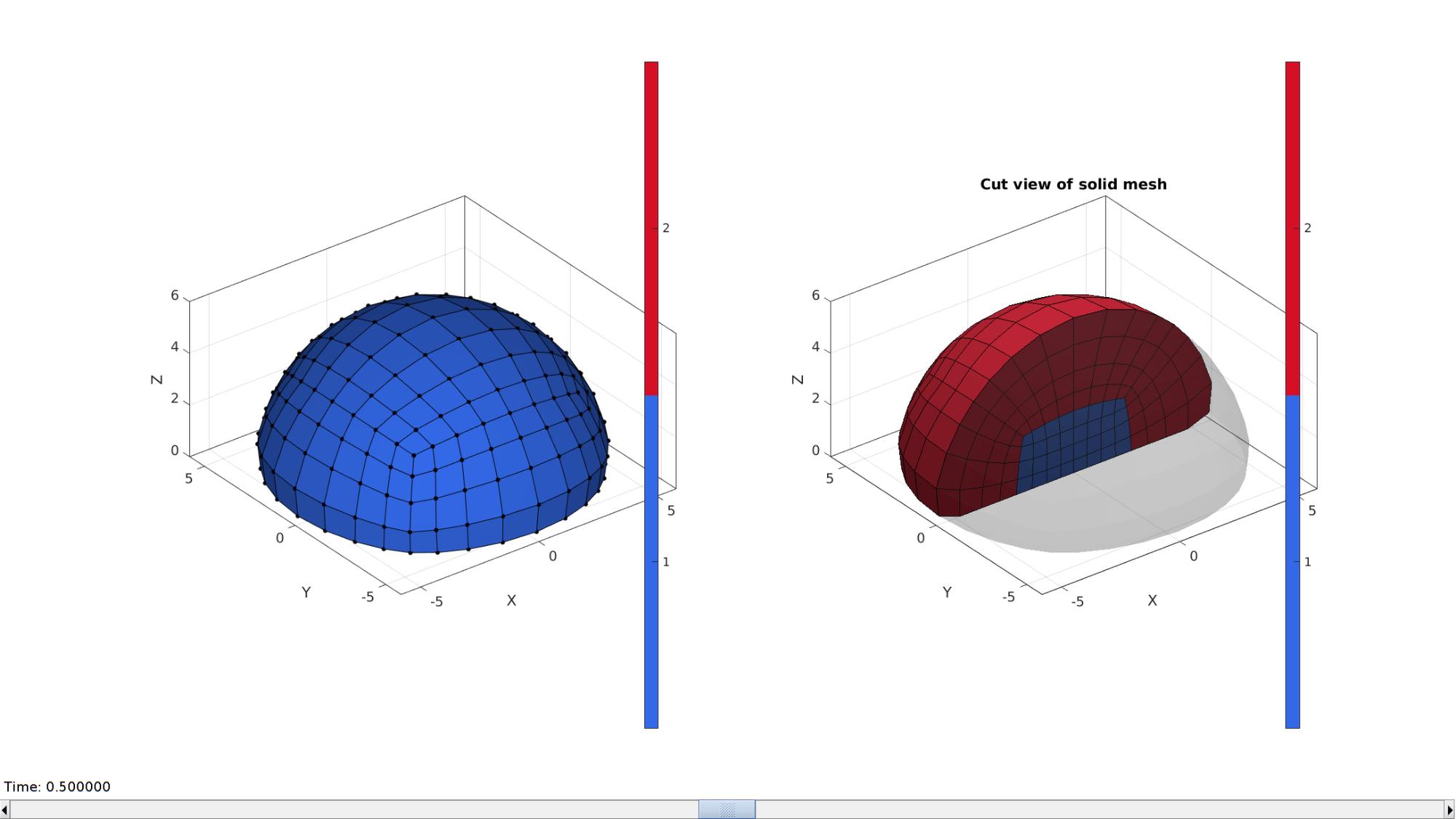
Visualize blob mesh
hFig=cFigure; subplot(1,2,1); hold on; hp=gpatch(Fb_blob_plot,V_blob,Cb_blob,'k',1); hp.Marker='.'; hp.MarkerSize=markerSize2; % patchNormPlot(Fb_blob,V_blob); % plotV(V_blob(indRigid,:),'g.','MarkerSize',25); axisGeom(gca,fontSize); colormap(gjet); icolorbar; camlight headlight; hs=subplot(1,2,2); hold on; title('Cut view of solid mesh','FontSize',fontSize); optionStruct.hFig=[hFig hs]; gpatch(Fb_blob_plot,V_blob,'kw','none',0.25); meshView(meshStruct,optionStruct); axisGeom(gca,fontSize); drawnow;
Creating rigid body shear surface
pointSpacingProbe=pointSpacingBlob/2; %Sketching side profile d=hemiSphereRadius*cos(asin(1-probeOverlapFactor)); x=[-probeLength-hemiSphereRadius -d -d]; y=[0 0 0]; z=[hemiSphereRadius*(1-probeOverlapFactor) hemiSphereRadius*(1-probeOverlapFactor) hemiSphereRadius*1.5]; V_probe_curve_sketch=[x(:) y(:) z(:)]; %Fillet sketch np=100; %Number of points used to construct each fillet edge [V_probe_curve]=filletCurve(V_probe_curve_sketch,filletProbe,np,0); numPointsProbeCurve=ceil(max(pathLength(V_probe_curve))/pointSpacingProbe); [V_probe_curve] = evenlySampleCurve(V_probe_curve,numPointsProbeCurve,'pchip',0); % Extruding curve % controlParametersExtrude.pointSpacing=pointSpacingProbe; controlParametersExtrude.depth=hemiSphereRadius*2.5; controlParametersExtrude.numSteps=ceil(controlParametersExtrude.depth/pointSpacingProbe); controlParametersExtrude.numSteps=controlParametersExtrude.numSteps+iseven(controlParametersExtrude.numSteps); %Force uneven controlParametersExtrude.patchType='quad'; controlParametersExtrude.dir=0; controlParametersExtrude.n=[0 1 0]; controlParametersExtrude.closeLoopOpt=0; [F_probe,V_probe]=polyExtrude(V_probe_curve,controlParametersExtrude); F_probe=fliplr(F_probe); %Invert face orientation so normals point to blob if strcmp(solidElementType,'hex20') [F_probe,V_probe]=quad4_quad8(F_probe,V_probe); [F_probe_plot]=element2patch(F_probe,[],'quad8'); else F_probe_plot=F_probe; end center_of_mass_probe=mean(V_probe,1);
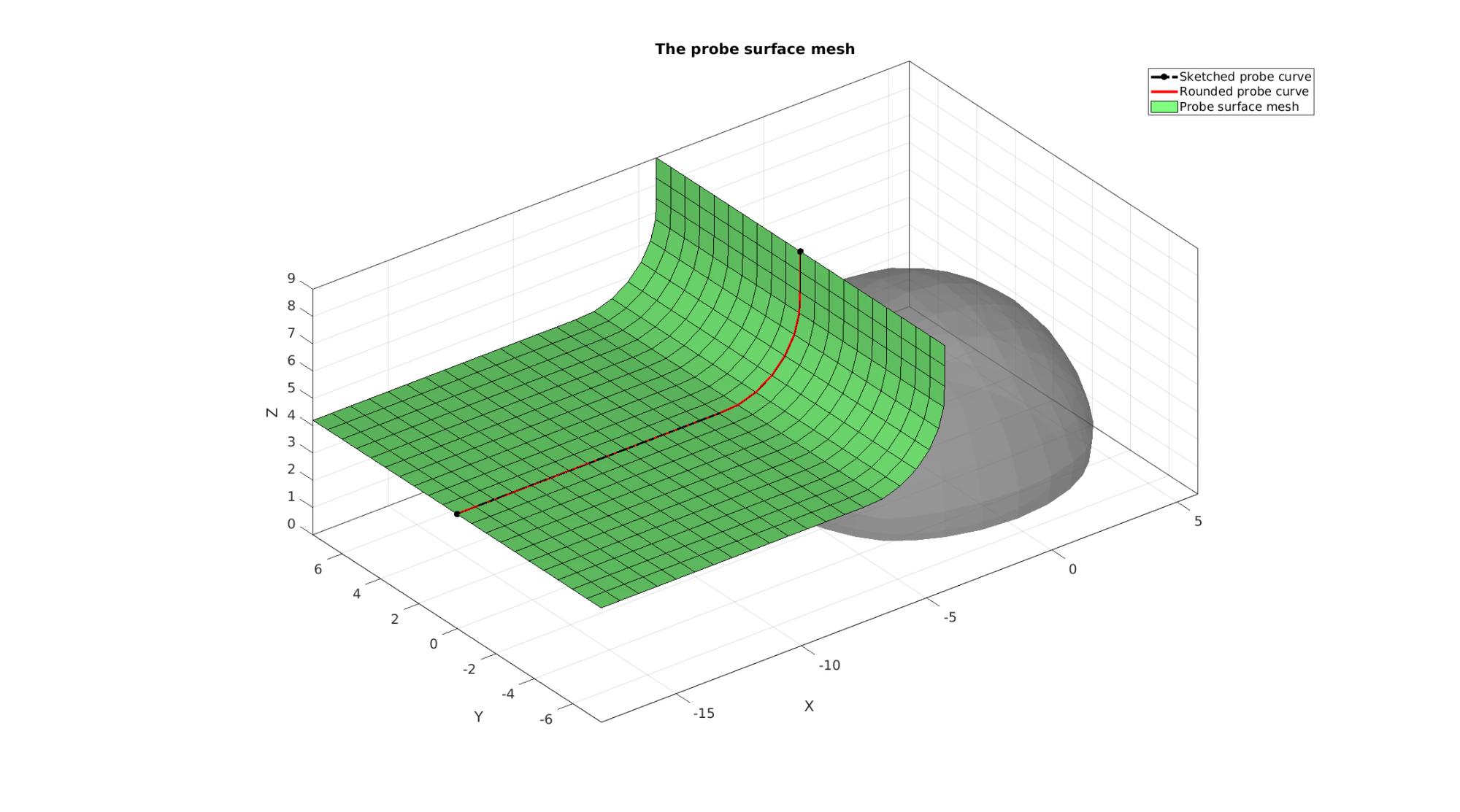
Visualizing probe mesh
cFigure; hold on; title('The probe surface mesh','fontSize',fontSize); gpatch(Fb_blob_plot,V_blob,'kw','none',0.5); hl(1)=plotV(V_probe_curve_sketch,'k.-.','lineWidth',3,'MarkerSize',25); hl(2)=plotV(V_probe_curve,'r-','lineWidth',3,'MarkerSize',25); hl(3)=gpatch(F_probe_plot,V_probe,'gw','k',1); % hl(3).Marker='.'; % hl(3).MarkerSize=markerSize2; legend(hl,{'Sketched probe curve','Rounded probe curve','Probe surface mesh'}); clear hl; axisGeom(gca,fontSize); camlight headlight; drawnow;
Join model node sets
V=[V_blob; V_probe]; F_probe=F_probe+size(V_blob,1); F_probe_plot=F_probe_plot+size(V_blob,1);
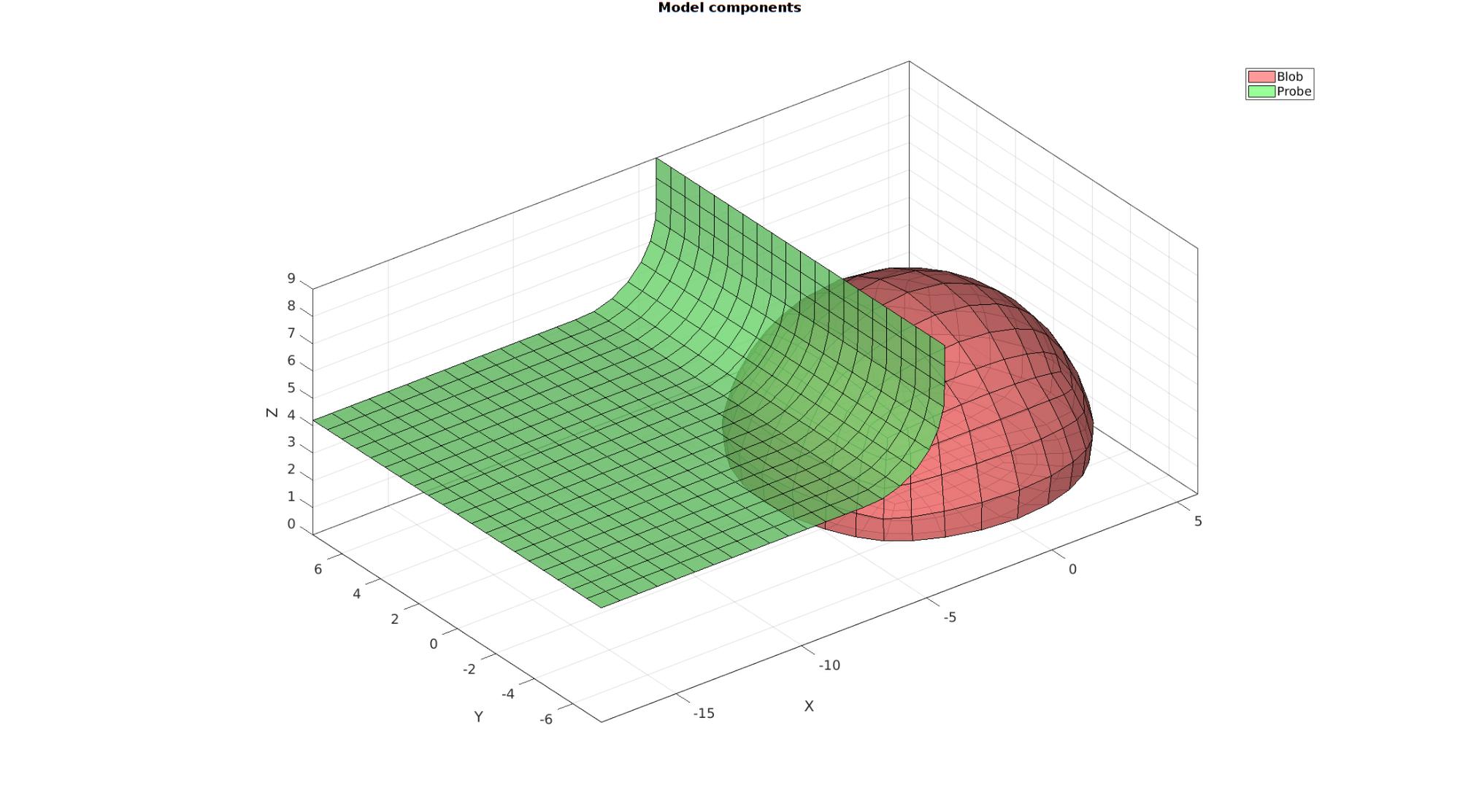
Visualizing model
cFigure; hold on; gtitle('Model components',fontSize); hl(1)=gpatch(Fb_blob_plot,V,'rw','k',0.8); hl(2)=gpatch(F_probe_plot,V,'gw','k',0.8); legend(hl,{'Blob','Probe'}); clear hl; axisGeom(gca,fontSize); camlight headlight; drawnow;
Get local material axes
[N3,Vn]=patchNormal(Fb_blob_plot,V);
[N1,N2]=vectorOrthogonalPair(N3); %Get orthogonal vector pair
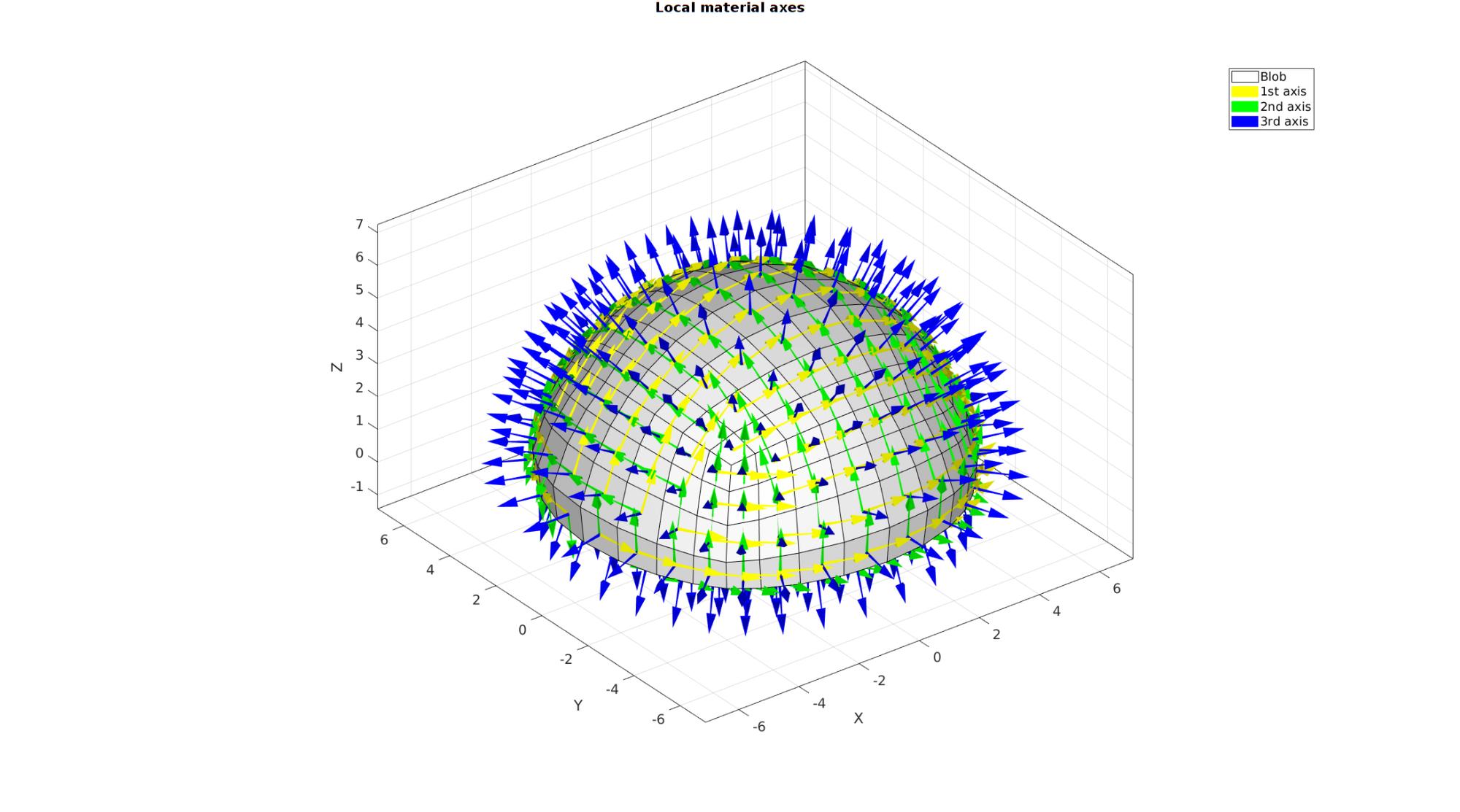
Visualizing axes
cFigure; hold on; gtitle('Local material axes',fontSize); hl(1)=gpatch(Fb_blob_plot,V,'w','k',1); hl(2)=quiverVec(Vn,N1,pointSpacingBlob,'y'); hl(3)=quiverVec(Vn,N2,pointSpacingBlob,'g'); hl(4)=quiverVec(Vn,N3,pointSpacingBlob,'b'); legend(hl,{'Blob','1st axis','2nd axis','3rd axis'}); clear hl; axisGeom(gca,fontSize); camlight headlight; drawnow;
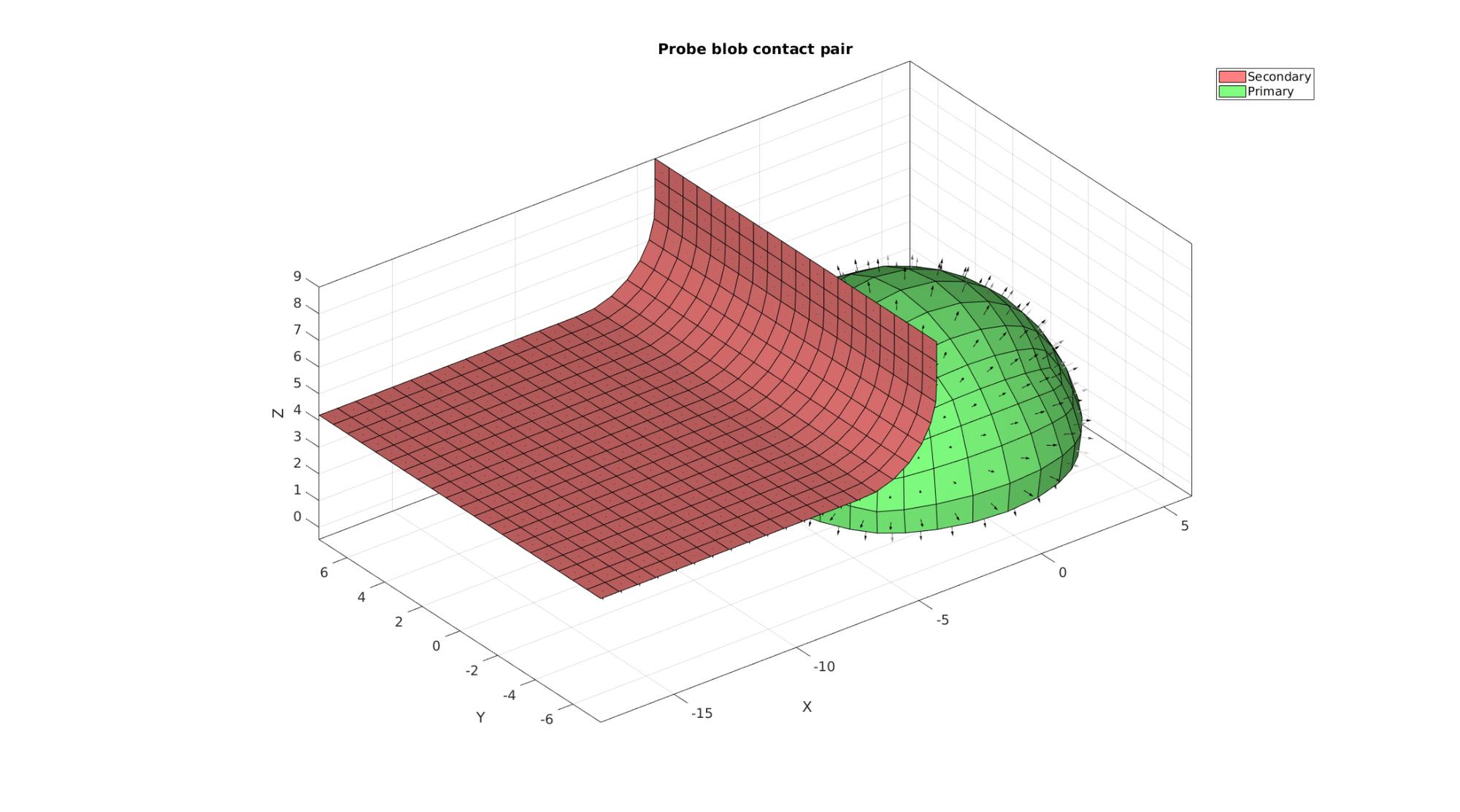
Visualize contact surfaces
cFigure; title('Probe blob contact pair','fontsize',fontSize); hl(1)=gpatch(F_probe_plot,V,'rw','k',1); patchNormPlot(F_probe_plot,V); hl(2)=gpatch(Fb_blob_plot,V,'gw','k',1); patchNormPlot(Fb_blob_plot,V); legend(hl,{'Secondary','Primary'}); clear hl; axisGeom(gca,fontSize); camlight headlight; drawnow;
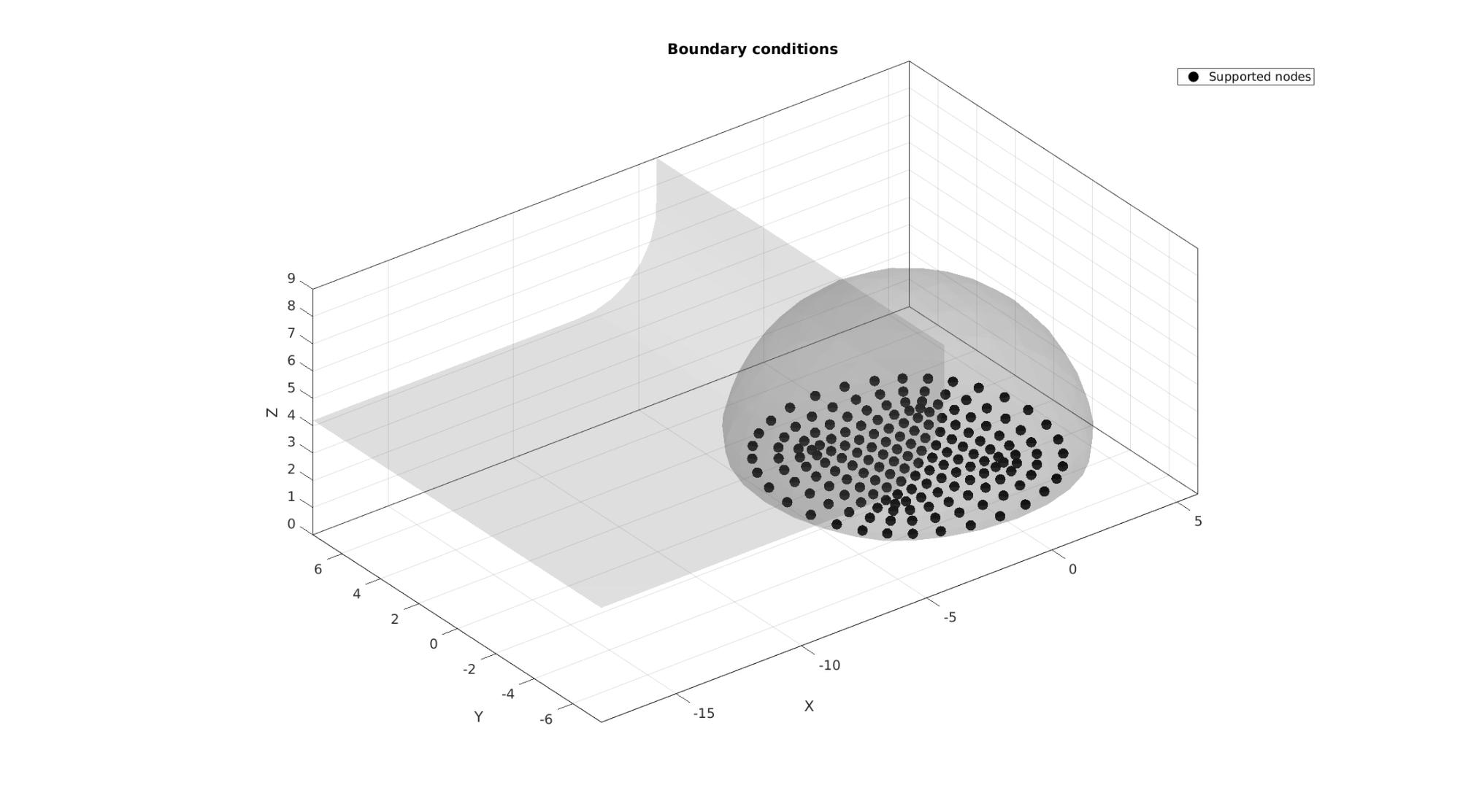
Setup boundary conditions
bcSupportList=unique(Fb_blob(Cb_blob==2,:));
Visualize boundary condition sets
cFigure; hold on; title('Boundary conditions ','fontsize',fontSize); gpatch(F_probe_plot,V,'kw','none',0.2); gpatch(Fb_blob_plot,V,'kw','none',0.2); hl(1)=plotV(V(bcSupportList,:),'k.','MarkerSize',markerSize); legend(hl,{'Supported nodes'}); clear hl; axisGeom(gca,fontSize); camlight headlight; drawnow;
Defining the FEBio input structure
See also febioStructTemplate and febioStruct2xml and the FEBio user manual.
%Get a template with default settings [febio_spec]=febioStructTemplate; %febio_spec version febio_spec.ATTR.version='4.0'; %Module section febio_spec.Module.ATTR.type='solid'; %Control section febio_spec.Control.analysis='STATIC'; febio_spec.Control.time_steps=numTimeSteps; febio_spec.Control.step_size=step_size; febio_spec.Control.solver.max_refs=max_refs; febio_spec.Control.solver.qn_method.max_ups=max_ups; febio_spec.Control.solver.symmetric_stiffness=symmetric_stiffness; febio_spec.Control.time_stepper.dtmin=dtmin; febio_spec.Control.time_stepper.dtmax=dtmax; febio_spec.Control.time_stepper.max_retries=max_retries; febio_spec.Control.time_stepper.opt_iter=opt_iter; %Material section materialName1='Material1'; febio_spec.Material.material{1}.ATTR.name=materialName1; febio_spec.Material.material{1}.ATTR.type='Ogden'; febio_spec.Material.material{1}.ATTR.id=1; febio_spec.Material.material{1}.c1=materialPropertiesOgden.c1; febio_spec.Material.material{1}.m1=materialPropertiesOgden.m1; febio_spec.Material.material{1}.c2=materialPropertiesOgden.c1; febio_spec.Material.material{1}.m2=-materialPropertiesOgden.m1; febio_spec.Material.material{1}.k=materialPropertiesOgden.k; materialName2='Material2'; febio_spec.Material.material{2}.ATTR.name=materialName2; febio_spec.Material.material{2}.ATTR.type='Fung orthotropic'; febio_spec.Material.material{2}.ATTR.id=2; febio_spec.Material.material{2}.E1=materialPropertiesFung.E1; febio_spec.Material.material{2}.E2=materialPropertiesFung.E2; febio_spec.Material.material{2}.E3=materialPropertiesFung.E3; febio_spec.Material.material{2}.G12=materialPropertiesFung.G12; febio_spec.Material.material{2}.G23=materialPropertiesFung.G23; febio_spec.Material.material{2}.G31=materialPropertiesFung.G31; febio_spec.Material.material{2}.v12=materialPropertiesFung.v12; febio_spec.Material.material{2}.v23=materialPropertiesFung.v23; febio_spec.Material.material{2}.v31=materialPropertiesFung.v31; febio_spec.Material.material{2}.c=materialPropertiesFung.c; febio_spec.Material.material{2}.k=materialPropertiesFung.k; materialName3='Material3'; febio_spec.Material.material{3}.ATTR.name=materialName3; febio_spec.Material.material{3}.ATTR.type='rigid body'; febio_spec.Material.material{3}.ATTR.id=3; febio_spec.Material.material{3}.density=1; febio_spec.Material.material{3}.center_of_mass=center_of_mass_probe; %Mesh section % -> Nodes febio_spec.Mesh.Nodes{1}.ATTR.name='nodeSet_all'; %The node set name febio_spec.Mesh.Nodes{1}.node.ATTR.id=(1:size(V,1))'; %The node id's febio_spec.Mesh.Nodes{1}.node.VAL=V; %The nodel coordinates % -> Elements partName1='Part1'; febio_spec.Mesh.Elements{1}.ATTR.name=partName1; %Name of this part febio_spec.Mesh.Elements{1}.ATTR.type=solidElementType; %Element type febio_spec.Mesh.Elements{1}.elem.ATTR.id=(1:1:size(E_blob,1))'; %Element id's febio_spec.Mesh.Elements{1}.elem.VAL=E_blob; %The element matrix partName2='Part2'; febio_spec.Mesh.Elements{2}.ATTR.name=partName2; %Name of this part febio_spec.Mesh.Elements{2}.ATTR.type=shellElementType; %Element type febio_spec.Mesh.Elements{2}.elem.ATTR.id=size(E_blob,1)+(1:1:size(Fb_blob,1))'; %Element id's febio_spec.Mesh.Elements{2}.elem.VAL=Fb_blob; partName3='Part3'; febio_spec.Mesh.Elements{3}.ATTR.name=partName3; %Name of this part febio_spec.Mesh.Elements{3}.ATTR.type=shellElementType; %Element type febio_spec.Mesh.Elements{3}.elem.ATTR.id=size(E_blob,1)+size(Fb_blob,1)+(1:1:size(F_probe,1))'; %Element id's febio_spec.Mesh.Elements{3}.elem.VAL=F_probe; % -> NodeSets nodeSetName1='bcSupportList'; febio_spec.Mesh.NodeSet{1}.ATTR.name=nodeSetName1; febio_spec.Mesh.NodeSet{1}.VAL=mrow(bcSupportList); %MeshDomains section febio_spec.MeshDomains.SolidDomain.ATTR.name=partName1; febio_spec.MeshDomains.SolidDomain.ATTR.mat=materialName1; febio_spec.MeshDomains.ShellDomain{1}.ATTR.name=partName2; febio_spec.MeshDomains.ShellDomain{1}.ATTR.mat=materialName2; febio_spec.MeshDomains.ShellDomain{1}.shell_thickness=membraneThickness; febio_spec.MeshDomains.ShellDomain{2}.ATTR.name=partName3; febio_spec.MeshDomains.ShellDomain{2}.ATTR.mat=materialName3; % % -> Surfaces surfaceName1='contactSurface1'; febio_spec.Mesh.Surface{1}.ATTR.name=surfaceName1; febio_spec.Mesh.Surface{1}.(shellElementType).ATTR.id=(1:1:size(F_probe,1))'; febio_spec.Mesh.Surface{1}.(shellElementType).VAL=F_probe; surfaceName2='contactSurface2'; febio_spec.Mesh.Surface{2}.ATTR.name=surfaceName2; febio_spec.Mesh.Surface{2}.(shellElementType).ATTR.id=(1:1:size(Fb_blob(Cb_blob==1,:),1))'; febio_spec.Mesh.Surface{2}.(shellElementType).VAL=Fb_blob(Cb_blob==1,:); % -> Surface pairs contactPairName1='Contact1'; febio_spec.Mesh.SurfacePair{1}.ATTR.name=contactPairName1; febio_spec.Mesh.SurfacePair{1}.primary=surfaceName2; febio_spec.Mesh.SurfacePair{1}.secondary=surfaceName1; %MeshData section % -> ElementData febio_spec.MeshData.ElementData{1}.ATTR.elem_set=partName2; febio_spec.MeshData.ElementData{1}.ATTR.type='mat_axis'; for q=1:1:size(N1,1) febio_spec.MeshData.ElementData{1}.elem{q}.ATTR.lid=q; febio_spec.MeshData.ElementData{1}.elem{q}.a=N1(q,:); febio_spec.MeshData.ElementData{1}.elem{q}.d=N2(q,:); end %Boundary condition section % -> Fix boundary conditions febio_spec.Boundary.bc{1}.ATTR.name='zero_displacement_xyz'; febio_spec.Boundary.bc{1}.ATTR.type='zero displacement'; febio_spec.Boundary.bc{1}.ATTR.node_set=nodeSetName1; febio_spec.Boundary.bc{1}.x_dof=1; febio_spec.Boundary.bc{1}.y_dof=1; febio_spec.Boundary.bc{1}.z_dof=1; %Rigid section % ->Rigid body fix boundary conditions febio_spec.Rigid.rigid_bc{1}.ATTR.name='RigidFix_1'; febio_spec.Rigid.rigid_bc{1}.ATTR.type='rigid_fixed'; febio_spec.Rigid.rigid_bc{1}.rb=3; % febio_spec.Rigid.rigid_bc{1}.Rx_dof=1; febio_spec.Rigid.rigid_bc{1}.Ry_dof=1; febio_spec.Rigid.rigid_bc{1}.Rz_dof=1; febio_spec.Rigid.rigid_bc{1}.Ru_dof=1; febio_spec.Rigid.rigid_bc{1}.Rv_dof=1; febio_spec.Rigid.rigid_bc{1}.Rw_dof=1; % ->Rigid body prescribe boundary conditions febio_spec.Rigid.rigid_bc{2}.ATTR.name='RigidPrescribe'; febio_spec.Rigid.rigid_bc{2}.ATTR.type='rigid_displacement'; febio_spec.Rigid.rigid_bc{2}.rb=3; febio_spec.Rigid.rigid_bc{2}.dof='x'; febio_spec.Rigid.rigid_bc{2}.value.ATTR.lc=1; febio_spec.Rigid.rigid_bc{2}.value.VAL=probeDisplacement; febio_spec.Rigid.rigid_bc{2}.relative=0; %Contact section febio_spec.Contact.contact{1}.ATTR.surface_pair=contactPairName1; febio_spec.Contact.contact{1}.ATTR.type='sliding-elastic'; febio_spec.Contact.contact{1}.two_pass=0; febio_spec.Contact.contact{1}.laugon=laugon; febio_spec.Contact.contact{1}.tolerance=0.2; febio_spec.Contact.contact{1}.gaptol=0; febio_spec.Contact.contact{1}.minaug=minaug; febio_spec.Contact.contact{1}.maxaug=maxaug; febio_spec.Contact.contact{1}.search_tol=0.01; febio_spec.Contact.contact{1}.search_radius=0.1*sqrt(sum((max(V,[],1)-min(V,[],1)).^2,2)); febio_spec.Contact.contact{1}.symmetric_stiffness=0; febio_spec.Contact.contact{1}.auto_penalty=1; febio_spec.Contact.contact{1}.penalty=contactPenalty; febio_spec.Contact.contact{1}.fric_coeff=fric_coeff; %LoadData section % -> load_controller febio_spec.LoadData.load_controller{1}.ATTR.name='LC_1'; febio_spec.LoadData.load_controller{1}.ATTR.id=1; febio_spec.LoadData.load_controller{1}.ATTR.type='loadcurve'; febio_spec.LoadData.load_controller{1}.interpolate='LINEAR'; %febio_spec.LoadData.load_controller{1}.extend='CONSTANT'; febio_spec.LoadData.load_controller{1}.points.pt.VAL=[0 0; 1 1]; %Output section % -> log file febio_spec.Output.logfile.ATTR.file=febioLogFileName; febio_spec.Output.logfile.node_data{1}.ATTR.file=febioLogFileName_disp; febio_spec.Output.logfile.node_data{1}.ATTR.data='ux;uy;uz'; febio_spec.Output.logfile.node_data{1}.ATTR.delim=','; % Plotfile section febio_spec.Output.plotfile.compression=0;
Quick viewing of the FEBio input file structure
The febView function can be used to view the xml structure in a MATLAB figure window.
febView(febio_spec); %Viewing the febio file
Exporting the FEBio input file
Exporting the febio_spec structure to an FEBio input file is done using the febioStruct2xml function.
febioStruct2xml(febio_spec,febioFebFileName); %Exporting to file and domNode
Running the FEBio analysis
To run the analysis defined by the created FEBio input file the runMonitorFEBio function is used. The input for this function is a structure defining job settings e.g. the FEBio input file name. The optional output runFlag informs the user if the analysis was run succesfully.
febioAnalysis.run_filename=febioFebFileName; %The input file name febioAnalysis.run_logname=febioLogFileName; %The name for the log file febioAnalysis.disp_on=1; %Display information on the command window febioAnalysis.runMode=runMode; [runFlag]=runMonitorFEBio(febioAnalysis);%START FEBio NOW!!!!!!!!
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
--------> RUNNING/MONITORING FEBIO JOB <-------- 20-Apr-2023 18:01:48
FEBio path: /home/kevin/FEBioStudio2/bin/febio4
# Attempt removal of existing log files 20-Apr-2023 18:01:48
* Removal succesful 20-Apr-2023 18:01:48
# Attempt removal of existing .xplt files 20-Apr-2023 18:01:48
* Removal succesful 20-Apr-2023 18:01:48
# Starting FEBio... 20-Apr-2023 18:01:48
Max. total analysis time is: Inf s
===========================================================================
________ _________ _______ __ _________
| |\ | |\ | \\ | |\ / \\
| ____|| | ____|| | __ || |__|| | ___ ||
| |\___\| | |\___\| | |\_| || \_\| | // \ ||
| ||__ | ||__ | ||_| || | |\ | || | ||
| |\ | |\ | \\ | || | || | ||
| ___|| | ___|| | ___ || | || | || | ||
| |\__\| | |\__\| | |\__| || | || | || | ||
| || | ||___ | ||__| || | || | \\__/ ||
| || | |\ | || | || | ||
|___|| |________|| |_________// |__|| \_________//
F I N I T E E L E M E N T S F O R B I O M E C H A N I C S
version 4.1.0
FEBio is a registered trademark.
copyright (c) 2006-2023 - All rights reserved
===========================================================================
Default linear solver: pardiso
Success loading plugin libFEBioChem.so (version 1.0.0)
Success loading plugin libFEBioHeat.so (version 2.0.0)
Reading file /mnt/data/MATLAB/GIBBON/data/temp/tempModel.feb ...SUCCESS!
Data Record #1
===========================================================================
Step = 0
Time = 0
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(0%) tempModel.feb - FEBio 4.1.0 ]0;(0%) tempModel.feb - FEBio 4.1.0
*************************************************************************
* Selecting linear solver pardiso *
*************************************************************************
===== beginning time step 1 : 0.04 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314749
1
Nonlinear solution status: time= 0.04
stiffness updates = 0
right hand side evaluations = 2
stiffness matrix reformations = 1
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.278678e-30 1.183365e-24 0.000000e+00
energy 1.249471e-31 1.700896e-32 1.249471e-33
displacement 3.812584e-30 3.812584e-30 3.812584e-36
*************************************************************************
* WARNING *
* No force acting on the system. *
*************************************************************************
convergence summary
number of iterations : 1
number of reformations : 1
------- converged at time : 0.04
Data Record #1
===========================================================================
Step = 1
Time = 0.04
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(4%) tempModel.feb - FEBio 4.1.0
===== beginning time step 2 : 0.08 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314749
1
Nonlinear solution status: time= 0.08
stiffness updates = 0
right hand side evaluations = 2
stiffness matrix reformations = 1
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.183365e-24 2.708415e-24 0.000000e+00
energy 2.982945e-28 1.819645e-28 2.982945e-30
displacement 2.484348e-29 2.484348e-29 2.484348e-35
*************************************************************************
* WARNING *
* No force acting on the system. *
*************************************************************************
convergence summary
number of iterations : 1
number of reformations : 1
------- converged at time : 0.08
Data Record #1
===========================================================================
Step = 2
Time = 0.08
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(8%) tempModel.feb - FEBio 4.1.0
===== beginning time step 3 : 0.12 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314749
1
Nonlinear solution status: time= 0.12
stiffness updates = 0
right hand side evaluations = 2
stiffness matrix reformations = 1
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.708415e-24 4.803897e-24 0.000000e+00
energy 6.727254e-28 4.301164e-28 6.727254e-30
displacement 7.859661e-29 7.859661e-29 7.859661e-35
*************************************************************************
* WARNING *
* No force acting on the system. *
*************************************************************************
convergence summary
number of iterations : 1
number of reformations : 1
------- converged at time : 0.12
Data Record #1
===========================================================================
Step = 3
Time = 0.12
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(12%) tempModel.feb - FEBio 4.1.0
===== beginning time step 4 : 0.16 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314749
1
Nonlinear solution status: time= 0.16
stiffness updates = 0
right hand side evaluations = 2
stiffness matrix reformations = 1
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.803897e-24 8.474256e-24 0.000000e+00
energy 1.351927e-27 9.715834e-28 1.351927e-29
displacement 1.645213e-28 1.645213e-28 1.645213e-34
*************************************************************************
* WARNING *
* No force acting on the system. *
*************************************************************************
convergence summary
number of iterations : 1
number of reformations : 1
------- converged at time : 0.16
Data Record #1
===========================================================================
Step = 4
Time = 0.16
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(16%) tempModel.feb - FEBio 4.1.0
===== beginning time step 5 : 0.2 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314749
1
Nonlinear solution status: time= 0.2
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 1
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 8.474256e-24 3.817631e-04 0.000000e+00
energy 2.106469e-27 7.292918e-18 2.106469e-29
displacement 2.597182e-28 6.492956e-29 6.492956e-35
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314839
2
Nonlinear solution status: time= 0.2
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 2
step from line search = 0.413601
convergence norms : INITIAL CURRENT REQUIRED
residual 3.817631e-04 3.236764e+00 0.000000e+00
energy 7.292918e-18 7.362321e-04 7.292918e-20
displacement 2.597182e-28 1.140667e-01 1.140667e-07
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314839
3
Nonlinear solution status: time= 0.2
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.236764e+00 8.726925e-01 0.000000e+00
energy 7.362321e-04 9.892783e-05 7.362321e-06
displacement 2.597182e-28 5.049775e-01 1.065808e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314803
4
Nonlinear solution status: time= 0.2
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.236764e+00 1.262363e+00 0.000000e+00
energy 7.362321e-04 1.423990e-04 7.362321e-06
displacement 2.597182e-28 2.171908e-02 9.774861e-07
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314803
5
Nonlinear solution status: time= 0.2
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.236764e+00 3.481272e-03 0.000000e+00
energy 7.362321e-04 1.354237e-05 7.362321e-06
displacement 2.597182e-28 3.816846e-03 9.210511e-07
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314803
6
Nonlinear solution status: time= 0.2
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.236764e+00 1.015401e-02 0.000000e+00
energy 7.362321e-04 8.194885e-07 7.362321e-06
displacement 2.597182e-28 2.749946e-03 8.848058e-07
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314803
7
Nonlinear solution status: time= 0.2
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.236764e+00 2.637022e-07 0.000000e+00
energy 7.362321e-04 1.710973e-08 7.362321e-06
displacement 2.597182e-28 1.530372e-05 8.822479e-07
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314803
8
Nonlinear solution status: time= 0.2
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.236764e+00 4.357759e-10 0.000000e+00
energy 7.362321e-04 3.493895e-12 7.362321e-06
displacement 2.597182e-28 3.636512e-07 8.819489e-07
convergence summary
number of iterations : 8
number of reformations : 8
------- converged at time : 0.2
Data Record #1
===========================================================================
Step = 5
Time = 0.2
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(20%) tempModel.feb - FEBio 4.1.0
===== beginning time step 6 : 0.24 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314803
1
Nonlinear solution status: time= 0.24
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 1
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.376855e-04 6.493037e+01 0.000000e+00
energy 1.138931e-02 5.157590e-03 1.138931e-04
displacement 2.145554e+00 5.363884e-01 5.363884e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
2
*************************************************************************
* ERROR *
* Negative jacobian was detected. *
*************************************************************************
------- failed to converge at time : 0.24
Retrying time step. Retry attempt 1 of max 25
AUTO STEPPER: retry step, dt = 0.0384615
===== beginning time step 6 : 0.238462 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314803
1
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 1
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 5.553508e+01 0.000000e+00
energy 1.053006e-02 4.112125e-03 1.053006e-04
displacement 1.983685e+00 4.959212e-01 4.959212e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
2
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 2
step from line search = 0.284680
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 2.069689e+01 0.000000e+00
energy 1.053006e-02 1.005679e-03 1.053006e-04
displacement 1.983685e+00 2.445595e-01 1.295697e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314857
3
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 3
step from line search = 0.385705
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 3.867870e+00 0.000000e+00
energy 1.053006e-02 9.402813e-04 1.053006e-04
displacement 1.983685e+00 9.785250e-02 1.843436e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314857
4
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 4
step from line search = 0.238391
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 3.235967e+00 0.000000e+00
energy 1.053006e-02 3.859329e-04 1.053006e-04
displacement 1.983685e+00 1.141388e-02 2.062877e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314857
5
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 13
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 1.935428e+00 0.000000e+00
energy 1.053006e-02 2.015541e-04 1.053006e-04
displacement 1.983685e+00 1.180272e-01 3.146953e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314857
6
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 14
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 3.730183e-01 0.000000e+00
energy 1.053006e-02 8.602314e-05 1.053006e-04
displacement 1.983685e+00 3.746821e-02 2.867910e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314857
7
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 15
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 9.328460e-02 0.000000e+00
energy 1.053006e-02 2.827550e-05 1.053006e-04
displacement 1.983685e+00 2.277573e-02 2.651628e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314827
8
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 2.821835e-02 0.000000e+00
energy 1.053006e-02 4.353802e-06 1.053006e-04
displacement 1.983685e+00 5.968287e-03 2.589910e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314827
9
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 17
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 2.121897e-04 0.000000e+00
energy 1.053006e-02 5.734087e-07 1.053006e-04
displacement 1.983685e+00 6.842545e-04 2.557646e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314827
10
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 18
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 2.318119e-06 0.000000e+00
energy 1.053006e-02 5.322452e-09 1.053006e-04
displacement 1.983685e+00 4.815197e-05 2.549294e-06
Reforming stiffness matrix: reformation #11
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314839
11
Nonlinear solution status: time= 0.238462
stiffness updates = 0
right hand side evaluations = 19
stiffness matrix reformations = 11
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.895885e-04 3.568393e-12 0.000000e+00
energy 1.053006e-02 5.918914e-12 1.053006e-04
displacement 1.983685e+00 6.939370e-08 2.548846e-06
convergence summary
number of iterations : 11
number of reformations : 11
------- converged at time : 0.238462
Data Record #1
===========================================================================
Step = 6
Time = 0.238461538
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(24%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0387196
===== beginning time step 7 : 0.277181 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314839
1
Nonlinear solution status: time= 0.277181
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 1
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.872507e-04 1.089111e+02 0.000000e+00
energy 1.725979e-02 1.131328e-02 1.725979e-04
displacement 2.817819e+00 7.044547e-01 7.044547e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314893
2
Nonlinear solution status: time= 0.277181
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 2
step from line search = 0.256890
convergence norms : INITIAL CURRENT REQUIRED
residual 9.872507e-04 4.137455e+01 0.000000e+00
energy 1.725979e-02 2.024316e-03 1.725979e-04
displacement 2.817819e+00 1.447334e-01 1.182591e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314893
3
Nonlinear solution status: time= 0.277181
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 3
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.872507e-04 4.290273e+04 0.000000e+00
energy 1.725979e-02 1.590528e+01 1.725979e-04
displacement 2.817819e+00 2.020738e+00 2.686956e-06
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
4
*************************************************************************
* ERROR *
* Negative jacobian was detected. *
*************************************************************************
------- failed to converge at time : 0.277181
Retrying time step. Retry attempt 1 of max 25
AUTO STEPPER: retry step, dt = 0.0372304
===== beginning time step 7 : 0.275692 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314839
1
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 1
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 9.316012e+01 0.000000e+00
energy 1.595764e-02 9.251666e-03 1.595764e-04
displacement 2.605231e+00 6.513078e-01 6.513078e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314893
2
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 2
step from line search = 0.328285
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 2.613463e+01 0.000000e+00
energy 1.595764e-02 1.594233e-03 1.595764e-04
displacement 2.605231e+00 2.309892e-01 1.332217e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314893
3
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 3
step from line search = 0.076856
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 1.572890e+01 0.000000e+00
energy 1.595764e-02 3.836825e-04 1.595764e-04
displacement 2.605231e+00 1.786203e-02 1.432005e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314893
4
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 14
stiffness matrix reformations = 4
step from line search = 0.596322
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 2.113108e+00 0.000000e+00
energy 1.595764e-02 6.107055e-04 1.595764e-04
displacement 2.605231e+00 3.566013e-01 2.887447e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314893
5
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 5
step from line search = 0.228326
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 2.061311e+00 0.000000e+00
energy 1.595764e-02 2.640298e-04 1.595764e-04
displacement 2.605231e+00 2.080235e-02 3.170363e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314893
6
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 18
stiffness matrix reformations = 6
step from line search = 0.604955
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 2.977479e+00 0.000000e+00
energy 1.595764e-02 1.756457e-04 1.595764e-04
displacement 2.605231e+00 2.037798e-02 3.405420e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
7
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 19
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 9.582721e-01 0.000000e+00
energy 1.595764e-02 1.168447e-04 1.595764e-04
displacement 2.605231e+00 2.782971e-02 3.539943e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
8
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 20
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 1.538488e-01 0.000000e+00
energy 1.595764e-02 6.016859e-05 1.595764e-04
displacement 2.605231e+00 2.833307e-02 3.313390e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
9
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 21
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 6.636953e-02 0.000000e+00
energy 1.595764e-02 1.447585e-05 1.595764e-04
displacement 2.605231e+00 1.463621e-02 3.209939e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
10
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 22
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 2.580757e-03 0.000000e+00
energy 1.595764e-02 3.046937e-06 1.595764e-04
displacement 2.605231e+00 2.137745e-03 3.184592e-06
Reforming stiffness matrix: reformation #11
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
11
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 23
stiffness matrix reformations = 11
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 7.268633e-05 0.000000e+00
energy 1.595764e-02 1.909758e-07 1.595764e-04
displacement 2.605231e+00 3.179347e-04 3.178457e-06
Reforming stiffness matrix: reformation #12
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
12
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 24
stiffness matrix reformations = 12
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 6.113022e-08 0.000000e+00
energy 1.595764e-02 3.289912e-09 1.595764e-04
displacement 2.605231e+00 6.483435e-06 3.179686e-06
Reforming stiffness matrix: reformation #13
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
13
Nonlinear solution status: time= 0.275692
stiffness updates = 0
right hand side evaluations = 25
stiffness matrix reformations = 13
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 9.127711e-04 4.931880e-12 0.000000e+00
energy 1.595764e-02 2.226969e-11 1.595764e-04
displacement 2.605231e+00 5.112098e-08 3.179956e-06
convergence summary
number of iterations : 13
number of reformations : 13
------- converged at time : 0.275692
Data Record #1
===========================================================================
Step = 7
Time = 0.275691936
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(28%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0374358
===== beginning time step 8 : 0.313128 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314869
1
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.277741
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 9.523094e+00 0.000000e+00
energy 2.691140e-02 5.314132e-03 2.691140e-04
displacement 3.523669e+00 2.718153e-01 2.718153e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314905
2
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 2.923484e+01 0.000000e+00
energy 2.691140e-02 3.734416e-03 2.691140e-04
displacement 3.523669e+00 3.165023e+00 5.042074e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
3
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.342609
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 1.176798e+01 0.000000e+00
energy 2.691140e-02 4.687076e-04 2.691140e-04
displacement 3.523669e+00 1.119750e-02 5.202345e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
4
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 4
step from line search = 0.490165
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 3.181133e+00 0.000000e+00
energy 2.691140e-02 2.676461e-04 2.691140e-04
displacement 3.523669e+00 1.274711e-02 5.237408e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
5
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 1.542501e+00 0.000000e+00
energy 2.691140e-02 1.178964e-04 2.691140e-04
displacement 3.523669e+00 9.460666e-02 4.647270e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
6
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 2.122366e-01 0.000000e+00
energy 2.691140e-02 7.180405e-05 2.691140e-04
displacement 3.523669e+00 5.803428e-02 4.212154e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
7
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 6.837760e-02 0.000000e+00
energy 2.691140e-02 1.583002e-05 2.691140e-04
displacement 3.523669e+00 2.922434e-02 3.968478e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
8
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 1.306928e-03 0.000000e+00
energy 2.691140e-02 2.161370e-06 2.691140e-04
displacement 3.523669e+00 3.722879e-03 3.899896e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
9
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 13
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 1.100552e-05 0.000000e+00
energy 2.691140e-02 5.497904e-08 2.691140e-04
displacement 3.523669e+00 3.089036e-04 3.882110e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
10
Nonlinear solution status: time= 0.313128
stiffness updates = 0
right hand side evaluations = 14
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.141729e-03 1.988286e-09 0.000000e+00
energy 2.691140e-02 5.495804e-10 2.691140e-04
displacement 3.523669e+00 1.994504e-06 3.881740e-06
convergence summary
number of iterations : 10
number of reformations : 10
------- converged at time : 0.313128
Data Record #1
===========================================================================
Step = 8
Time = 0.31312776
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(31%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0379487
===== beginning time step 9 : 0.351076 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
1
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.284673
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 1.509694e+01 0.000000e+00
energy 3.767541e-02 8.957060e-03 3.767541e-04
displacement 4.649691e+00 3.768062e-01 3.768062e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314905
2
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 2.285717e+01 0.000000e+00
energy 3.767541e-02 2.691156e-03 3.767541e-04
displacement 4.649691e+00 4.544457e+00 7.122779e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
3
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 3
step from line search = 0.030803
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 2.234447e+01 0.000000e+00
energy 3.767541e-02 2.226000e-04 3.767541e-04
displacement 4.649691e+00 1.676038e-02 6.751514e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
4
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 2.074515e+01 0.000000e+00
energy 3.767541e-02 3.804894e-03 3.767541e-04
displacement 4.649691e+00 1.080896e-01 6.796579e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
5
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 1.441255e+00 0.000000e+00
energy 3.767541e-02 7.688207e-04 3.767541e-04
displacement 4.649691e+00 2.312281e-01 5.684720e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
6
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 6
step from line search = 0.081303
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 1.171558e+00 0.000000e+00
energy 3.767541e-02 9.384495e-05 3.767541e-04
displacement 4.649691e+00 4.508402e-03 5.637466e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
7
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 13
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 2.362039e+00 0.000000e+00
energy 3.767541e-02 1.858437e-04 3.767541e-04
displacement 4.649691e+00 1.110780e-01 5.290259e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
8
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 14
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 2.821331e-02 0.000000e+00
energy 3.767541e-02 4.086481e-05 3.767541e-04
displacement 4.649691e+00 2.001208e-02 5.120771e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
9
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 15
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 8.905704e-02 0.000000e+00
energy 3.767541e-02 2.062700e-06 3.767541e-04
displacement 4.649691e+00 2.695327e-02 5.022686e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
10
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 1.576938e-04 0.000000e+00
energy 3.767541e-02 1.141977e-06 3.767541e-04
displacement 4.649691e+00 1.612968e-03 4.983352e-06
Reforming stiffness matrix: reformation #11
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
11
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 17
stiffness matrix reformations = 11
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 1.575564e-05 0.000000e+00
energy 3.767541e-02 3.857916e-08 3.767541e-04
displacement 4.649691e+00 2.757249e-04 4.972620e-06
Reforming stiffness matrix: reformation #12
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
12
Nonlinear solution status: time= 0.351076
stiffness updates = 0
right hand side evaluations = 18
stiffness matrix reformations = 12
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.362062e-03 5.948709e-10 0.000000e+00
energy 3.767541e-02 8.366199e-10 3.767541e-04
displacement 4.649691e+00 4.788945e-06 4.969651e-06
convergence summary
number of iterations : 12
number of reformations : 12
------- converged at time : 0.351076
Data Record #1
===========================================================================
Step = 9
Time = 0.35107642
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(35%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0381908
===== beginning time step 10 : 0.389267 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314875
1
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 1
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 8.349732e+01 0.000000e+00
energy 2.916982e-02 3.526637e-04 2.916982e-04
displacement 4.074143e+00 1.018536e+00 1.018536e-06
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314953
2
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 2
step from line search = 0.085634
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 6.388132e+01 0.000000e+00
energy 2.916982e-02 1.618364e-03 2.916982e-04
displacement 4.074143e+00 1.658491e-02 1.167422e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314953
3
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 3
step from line search = 0.383778
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 1.696115e+01 0.000000e+00
energy 2.916982e-02 2.366214e-03 2.916982e-04
displacement 4.074143e+00 2.683168e-01 2.318475e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314953
4
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 1.965652e+01 0.000000e+00
energy 2.916982e-02 4.778094e-03 2.916982e-04
displacement 4.074143e+00 8.738353e-01 5.496527e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
5
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 13
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 4.603624e+00 0.000000e+00
energy 2.916982e-02 5.189865e-04 2.916982e-04
displacement 4.074143e+00 1.297666e-01 5.003764e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
6
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 14
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 1.493884e+00 0.000000e+00
energy 2.916982e-02 1.002689e-04 2.916982e-04
displacement 4.074143e+00 1.646089e-01 4.605392e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
7
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 15
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 3.652673e-01 0.000000e+00
energy 2.916982e-02 6.052162e-05 2.916982e-04
displacement 4.074143e+00 5.556393e-02 4.573065e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
8
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 6.592100e-02 0.000000e+00
energy 2.916982e-02 2.666669e-05 2.916982e-04
displacement 4.074143e+00 3.023559e-02 4.610790e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
9
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 17
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 6.864901e-03 0.000000e+00
energy 2.916982e-02 5.702066e-06 2.916982e-04
displacement 4.074143e+00 7.166961e-03 4.665974e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
10
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 18
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 1.332411e-04 0.000000e+00
energy 2.916982e-02 5.642941e-07 2.916982e-04
displacement 4.074143e+00 8.987634e-04 4.698566e-06
Reforming stiffness matrix: reformation #11
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
11
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 19
stiffness matrix reformations = 11
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 3.884122e-07 0.000000e+00
energy 2.916982e-02 2.465383e-08 2.916982e-04
displacement 4.074143e+00 3.263109e-05 4.709530e-06
Reforming stiffness matrix: reformation #12
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
12
Nonlinear solution status: time= 0.389267
stiffness updates = 0
right hand side evaluations = 20
stiffness matrix reformations = 12
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.552145e-03 2.038845e-10 0.000000e+00
energy 2.916982e-02 4.831561e-10 2.916982e-04
displacement 4.074143e+00 5.337405e-07 4.711566e-06
convergence summary
number of iterations : 12
number of reformations : 12
------- converged at time : 0.389267
Data Record #1
===========================================================================
Step = 10
Time = 0.389267208
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(39%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0384043
===== beginning time step 11 : 0.427672 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
1
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.365549
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 2.691370e+01 0.000000e+00
energy 4.187564e-02 9.085216e-03 4.187564e-04
displacement 4.017157e+00 5.367979e-01 5.367979e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314965
2
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 7.779857e+00 0.000000e+00
energy 4.187564e-02 1.231201e-03 4.187564e-04
displacement 4.017157e+00 2.571250e+00 5.110339e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314941
3
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.320000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 5.248766e+00 0.000000e+00
energy 4.187564e-02 5.877189e-04 4.187564e-04
displacement 4.017157e+00 7.427427e-02 4.551859e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314959
4
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 1.754771e+00 0.000000e+00
energy 4.187564e-02 5.842658e-05 4.187564e-04
displacement 4.017157e+00 4.415552e-02 4.484993e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314959
5
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 4.829240e-01 0.000000e+00
energy 4.187564e-02 6.136205e-05 4.187564e-04
displacement 4.017157e+00 1.764777e-01 3.893392e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314959
6
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 9.949356e-02 0.000000e+00
energy 4.187564e-02 2.635601e-05 4.187564e-04
displacement 4.017157e+00 3.585680e-02 3.841135e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
7
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 5.554549e-03 0.000000e+00
energy 4.187564e-02 5.655133e-06 4.187564e-04
displacement 4.017157e+00 1.342412e-02 3.799016e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
8
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 1.368139e-04 0.000000e+00
energy 4.187564e-02 5.089791e-07 4.187564e-04
displacement 4.017157e+00 1.464757e-03 3.805016e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
9
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 2.679603e-07 0.000000e+00
energy 4.187564e-02 2.202819e-08 4.187564e-04
displacement 4.017157e+00 5.944012e-05 3.808090e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
10
Nonlinear solution status: time= 0.427672
stiffness updates = 0
right hand side evaluations = 13
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.736372e-03 2.218556e-10 0.000000e+00
energy 4.187564e-02 5.947319e-10 4.187564e-04
displacement 4.017157e+00 1.388205e-06 3.808864e-06
convergence summary
number of iterations : 10
number of reformations : 10
------- converged at time : 0.427672
Data Record #1
===========================================================================
Step = 11
Time = 0.427671544
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(43%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0387235
===== beginning time step 12 : 0.466395 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
1
Nonlinear solution status: time= 0.466395
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.390670
convergence norms : INITIAL CURRENT REQUIRED
residual 8.196481e-03 3.950702e+01 0.000000e+00
energy 5.317277e-02 1.332794e-02 5.317277e-04
displacement 4.916969e+00 7.504411e-01 7.504411e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315013
2
Nonlinear solution status: time= 0.466395
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 8.196481e-03 8.578087e+01 0.000000e+00
energy 5.317277e-02 2.423460e-02 5.317277e-04
displacement 4.916969e+00 2.452162e+00 5.716351e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
3
Nonlinear solution status: time= 0.466395
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.138025
convergence norms : INITIAL CURRENT REQUIRED
residual 8.196481e-03 5.493847e+01 0.000000e+00
energy 5.317277e-02 1.127436e-03 5.317277e-04
displacement 4.916969e+00 2.604055e-02 6.083769e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
4
Nonlinear solution status: time= 0.466395
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 4
step from line search = 0.086309
convergence norms : INITIAL CURRENT REQUIRED
residual 8.196481e-03 4.289152e+01 0.000000e+00
energy 5.317277e-02 5.877111e-04 5.317277e-04
displacement 4.916969e+00 1.921623e-02 6.486131e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
5
Nonlinear solution status: time= 0.466395
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 5
step from line search = 0.097028
convergence norms : INITIAL CURRENT REQUIRED
residual 8.196481e-03 3.059716e+01 0.000000e+00
energy 5.317277e-02 1.053073e-03 5.317277e-04
displacement 4.916969e+00 1.286099e-02 6.481884e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
6
Nonlinear solution status: time= 0.466395
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 6
step from line search = 0.062581
convergence norms : INITIAL CURRENT REQUIRED
residual 8.196481e-03 2.569443e+01 0.000000e+00
energy 5.317277e-02 1.355969e-04 5.317277e-04
displacement 4.916969e+00 6.048984e-03 6.717688e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
7
*************************************************************************
* ERROR *
* Negative jacobian was detected. *
*************************************************************************
------- failed to converge at time : 0.466395
Retrying time step. Retry attempt 1 of max 25
AUTO STEPPER: retry step, dt = 0.0372341
===== beginning time step 12 : 0.464906 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
1
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.403139
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 3.827099e+01 0.000000e+00
energy 4.916129e-02 1.223144e-02 4.916129e-04
displacement 4.546009e+00 7.388221e-01 7.388221e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315013
2
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 5.637274e+01 0.000000e+00
energy 4.916129e-02 1.671952e-02 4.916129e-04
displacement 4.546009e+00 2.311880e+00 5.457305e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
3
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.135009
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 3.865632e+01 0.000000e+00
energy 4.916129e-02 8.046051e-04 4.916129e-04
displacement 4.546009e+00 2.317566e-02 5.889256e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
4
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 4
step from line search = 0.198897
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 2.024995e+01 0.000000e+00
energy 4.916129e-02 9.901053e-04 4.916129e-04
displacement 4.546009e+00 1.922051e-02 6.178665e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
5
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 5
step from line search = 0.058133
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 1.583045e+01 0.000000e+00
energy 4.916129e-02 5.434144e-04 4.916129e-04
displacement 4.546009e+00 6.443682e-03 6.238255e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
6
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 6
step from line search = 0.081290
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 1.227298e+01 0.000000e+00
energy 4.916129e-02 1.330745e-04 4.916129e-04
displacement 4.546009e+00 7.505053e-03 6.542766e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
7
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 7
step from line search = 0.061784
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 8.368614e+00 0.000000e+00
energy 4.916129e-02 2.276272e-05 4.916129e-04
displacement 4.546009e+00 1.079256e-02 6.853723e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
8
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 20
stiffness matrix reformations = 8
step from line search = 0.032978
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 6.789802e+00 0.000000e+00
energy 4.916129e-02 7.781463e-05 4.916129e-04
displacement 4.546009e+00 3.310319e-03 6.915857e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
9
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 23
stiffness matrix reformations = 9
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 7.578172e-03 3.134340e+04 0.000000e+00
energy 4.916129e-02 6.919573e+00 4.916129e-04
displacement 4.546009e+00 5.307529e+00 1.785083e-05
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314959
10
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 25
stiffness matrix reformations = 10
step from line search = 0.423851
convergence norms : INITIAL CURRENT REQUIRED
residual 3.134340e+04 7.238315e+03 0.000000e+00
energy 6.919573e+00 5.352300e-01 6.919573e-02
displacement 4.546009e+00 3.326799e+00 2.567253e-05
Reforming stiffness matrix: reformation #11
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314947
11
Nonlinear solution status: time= 0.464906
stiffness updates = 0
right hand side evaluations = 27
stiffness matrix reformations = 11
step from line search = 0.506017
convergence norms : INITIAL CURRENT REQUIRED
residual 3.134340e+04 1.853575e+03 0.000000e+00
energy 6.919573e+00 1.258319e-01 6.919573e-02
displacement 4.546009e+00 1.877006e+00 2.821579e-05
Reforming stiffness matrix: reformation #12
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314947
12
*************************************************************************
* ERROR *
* Negative jacobian was detected. *
*************************************************************************
------- failed to converge at time : 0.464906
Retrying time step. Retry attempt 2 of max 25
AUTO STEPPER: retry step, dt = 0.0357447
===== beginning time step 12 : 0.463416 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
1
Nonlinear solution status: time= 0.463416
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.416514
convergence norms : INITIAL CURRENT REQUIRED
residual 6.984107e-03 3.701635e+01 0.000000e+00
energy 4.530711e-02 1.111823e-02 4.530711e-04
displacement 4.189597e+00 7.268262e-01 7.268262e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315013
2
Nonlinear solution status: time= 0.463416
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.984107e-03 3.698964e+01 0.000000e+00
energy 4.530711e-02 1.087076e-02 4.530711e-04
displacement 4.189597e+00 2.249128e+00 5.305355e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
3
Nonlinear solution status: time= 0.463416
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 3
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.984107e-03 1.421607e+05 0.000000e+00
energy 4.530711e-02 2.807299e+01 4.530711e-04
displacement 4.189597e+00 7.706838e+00 9.120211e-06
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
4
*************************************************************************
* ERROR *
* Negative jacobian was detected. *
*************************************************************************
------- failed to converge at time : 0.463416
Retrying time step. Retry attempt 3 of max 25
AUTO STEPPER: retry step, dt = 0.0342554
===== beginning time step 12 : 0.461927 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
1
Nonlinear solution status: time= 0.461927
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.430744
convergence norms : INITIAL CURRENT REQUIRED
residual 6.414288e-03 3.569043e+01 0.000000e+00
energy 4.161025e-02 9.994130e-03 4.161025e-04
displacement 3.847733e+00 7.139101e-01 7.139101e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315013
2
Nonlinear solution status: time= 0.461927
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.414288e-03 2.417102e+01 0.000000e+00
energy 4.161025e-02 6.804796e-03 4.161025e-04
displacement 3.847733e+00 2.194165e+00 5.152393e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
3
Nonlinear solution status: time= 0.461927
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 3
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.414288e-03 7.691201e+04 0.000000e+00
energy 4.161025e-02 1.778246e+01 4.161025e-04
displacement 3.847733e+00 6.385700e+00 1.510840e-05
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314965
4
*************************************************************************
* ERROR *
* Negative jacobian was detected. *
*************************************************************************
------- failed to converge at time : 0.461927
Retrying time step. Retry attempt 4 of max 25
AUTO STEPPER: retry step, dt = 0.032766
===== beginning time step 12 : 0.460438 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
1
Nonlinear solution status: time= 0.460438
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.445939
convergence norms : INITIAL CURRENT REQUIRED
residual 5.868713e-03 3.430102e+01 0.000000e+00
energy 3.807069e-02 8.940162e-03 3.807069e-04
displacement 3.520416e+00 7.000771e-01 7.000771e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315001
2
Nonlinear solution status: time= 0.460438
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.868713e-03 1.691555e+01 0.000000e+00
energy 3.807069e-02 4.983523e-03 3.807069e-04
displacement 3.520416e+00 2.012769e+00 4.840882e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
3
Nonlinear solution status: time= 0.460438
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.195285
convergence norms : INITIAL CURRENT REQUIRED
residual 5.868713e-03 7.403208e+00 0.000000e+00
energy 3.807069e-02 4.551004e-04 3.807069e-04
displacement 3.520416e+00 2.977858e-02 5.280078e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
4
Nonlinear solution status: time= 0.460438
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 4
step from line search = 0.028937
convergence norms : INITIAL CURRENT REQUIRED
residual 5.868713e-03 4.924372e+00 0.000000e+00
energy 3.807069e-02 1.757260e-04 3.807069e-04
displacement 3.520416e+00 1.008203e-02 5.232897e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
5
Nonlinear solution status: time= 0.460438
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 5
step from line search = 0.048305
convergence norms : INITIAL CURRENT REQUIRED
residual 5.868713e-03 3.930388e+00 0.000000e+00
energy 3.807069e-02 2.028461e-04 3.807069e-04
displacement 3.520416e+00 7.344948e-03 5.368483e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
6
Nonlinear solution status: time= 0.460438
stiffness updates = 0
right hand side evaluations = 15
stiffness matrix reformations = 6
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.868713e-03 3.061025e+03 0.000000e+00
energy 3.807069e-02 5.443839e-01 3.807069e-04
displacement 3.520416e+00 2.555916e+00 1.316123e-05
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
7
Nonlinear solution status: time= 0.460438
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.061025e+03 9.753870e+00 0.000000e+00
energy 5.443839e-01 2.785067e-02 5.443839e-03
displacement 3.520416e+00 6.411978e-01 1.342625e-05
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314995
8
*************************************************************************
* ERROR *
* Negative jacobian was detected. *
*************************************************************************
------- failed to converge at time : 0.460438
Retrying time step. Retry attempt 5 of max 25
AUTO STEPPER: retry step, dt = 0.0312766
===== beginning time step 12 : 0.458948 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
1
Nonlinear solution status: time= 0.458948
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.462284
convergence norms : INITIAL CURRENT REQUIRED
residual 5.347383e-03 3.287230e+01 0.000000e+00
energy 3.468845e-02 8.062243e-03 3.468845e-04
displacement 3.207647e+00 6.854956e-01 6.854956e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
2
Nonlinear solution status: time= 0.458948
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.347383e-03 2.265419e+01 0.000000e+00
energy 3.468845e-02 1.214921e-02 3.468845e-04
displacement 3.207647e+00 1.723671e+00 4.217365e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
3
Nonlinear solution status: time= 0.458948
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.079361
convergence norms : INITIAL CURRENT REQUIRED
residual 5.347383e-03 1.841134e+01 0.000000e+00
energy 3.468845e-02 3.371844e-04 3.468845e-04
displacement 3.207647e+00 9.847705e-03 4.337047e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
4
Nonlinear solution status: time= 0.458948
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 4
step from line search = 0.129600
convergence norms : INITIAL CURRENT REQUIRED
residual 5.347383e-03 1.396830e+01 0.000000e+00
energy 3.468845e-02 5.956021e-04 3.468845e-04
displacement 3.207647e+00 7.343255e-03 4.457185e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
5
Nonlinear solution status: time= 0.458948
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 5
step from line search = 0.065458
convergence norms : INITIAL CURRENT REQUIRED
residual 5.347383e-03 1.458517e+01 0.000000e+00
energy 3.468845e-02 3.851808e-04 3.468845e-04
displacement 3.207647e+00 2.478920e-02 4.331372e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
6
Nonlinear solution status: time= 0.458948
stiffness updates = 0
right hand side evaluations = 14
stiffness matrix reformations = 6
step from line search = 0.493171
convergence norms : INITIAL CURRENT REQUIRED
residual 5.347383e-03 2.375037e+00 0.000000e+00
energy 3.468845e-02 2.636381e-04 3.468845e-04
displacement 3.207647e+00 1.088401e-01 5.160052e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
7
Nonlinear solution status: time= 0.458948
stiffness updates = 0
right hand side evaluations = 17
stiffness matrix reformations = 7
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.347383e-03 1.000984e+05 0.000000e+00
energy 3.468845e-02 2.220995e+01 3.468845e-04
displacement 3.207647e+00 1.425944e+01 3.006985e-05
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314995
8
*************************************************************************
* ERROR *
* Negative jacobian was detected. *
*************************************************************************
------- failed to converge at time : 0.458948
Retrying time step. Retry attempt 6 of max 25
AUTO STEPPER: retry step, dt = 0.0297873
===== beginning time step 12 : 0.457459 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314971
1
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.479860
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 3.138660e+01 0.000000e+00
energy 3.146351e-02 7.315219e-03 3.146351e-04
displacement 2.909426e+00 6.699412e-01 6.699412e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
2
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 1.443708e+01 0.000000e+00
energy 3.146351e-02 8.067828e-03 3.146351e-04
displacement 2.909426e+00 1.553419e+00 3.944498e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
3
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.213830
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 8.781366e+00 0.000000e+00
energy 3.146351e-02 5.341241e-04 3.146351e-04
displacement 2.909426e+00 2.015374e-02 4.192678e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
4
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 4
step from line search = 0.065123
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 5.423019e+00 0.000000e+00
energy 3.146351e-02 1.658281e-04 3.146351e-04
displacement 2.909426e+00 9.192922e-03 4.169818e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
5
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 5
step from line search = 0.156713
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 3.743097e+00 0.000000e+00
energy 3.146351e-02 4.788846e-04 3.146351e-04
displacement 2.909426e+00 8.908284e-03 4.323404e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
6
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 6
step from line search = 0.066397
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 5.113220e+00 0.000000e+00
energy 3.146351e-02 3.097844e-04 3.146351e-04
displacement 2.909426e+00 5.286576e-02 3.846460e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
7
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 17
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 4.385452e+00 0.000000e+00
energy 3.146351e-02 5.843413e-04 3.146351e-04
displacement 2.909426e+00 3.757694e-02 4.038988e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
8
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 18
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 5.664929e-01 0.000000e+00
energy 3.146351e-02 1.899295e-05 3.146351e-04
displacement 2.909426e+00 1.021694e-01 3.712786e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
9
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 19
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 3.485761e-01 0.000000e+00
energy 3.146351e-02 1.264306e-05 3.146351e-04
displacement 2.909426e+00 5.548728e-02 3.792428e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
10
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 20
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 1.565178e-03 0.000000e+00
energy 3.146351e-02 4.648547e-06 3.146351e-04
displacement 2.909426e+00 7.540670e-03 3.829782e-06
Reforming stiffness matrix: reformation #11
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
11
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 21
stiffness matrix reformations = 11
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 3.554838e-04 0.000000e+00
energy 3.146351e-02 3.295764e-07 3.146351e-04
displacement 2.909426e+00 2.317376e-03 3.844623e-06
Reforming stiffness matrix: reformation #12
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
12
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 22
stiffness matrix reformations = 12
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 8.340966e-08 0.000000e+00
energy 3.146351e-02 1.506437e-08 3.146351e-04
displacement 2.909426e+00 1.015800e-04 3.848531e-06
Reforming stiffness matrix: reformation #13
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
13
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 23
stiffness matrix reformations = 13
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 9.384315e-11 0.000000e+00
energy 3.146351e-02 5.583620e-10 3.146351e-04
displacement 2.909426e+00 3.945681e-06 3.849296e-06
Reforming stiffness matrix: reformation #14
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
14
Nonlinear solution status: time= 0.457459
stiffness updates = 0
right hand side evaluations = 24
stiffness matrix reformations = 14
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.850298e-03 2.834648e-13 0.000000e+00
energy 3.146351e-02 2.007491e-11 3.146351e-04
displacement 2.909426e+00 1.401372e-07 3.849440e-06
convergence summary
number of iterations : 14
number of reformations : 14
------- converged at time : 0.457459
Data Record #1
===========================================================================
Step = 12
Time = 0.457458828
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(46%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0301457
===== beginning time step 13 : 0.487605 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
1
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.490021
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 2.303364e+01 0.000000e+00
energy 2.600268e-02 3.943458e-03 2.600268e-04
displacement 2.415224e+00 5.799438e-01 5.799438e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
2
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 9.032950e-01 0.000000e+00
energy 2.600268e-02 3.170998e-04 2.600268e-04
displacement 2.415224e+00 4.458467e-01 1.895625e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
3
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.563292
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 1.935504e+00 0.000000e+00
energy 2.600268e-02 1.476902e-04 2.600268e-04
displacement 2.415224e+00 8.900371e-02 1.797342e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
4
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 2.575898e-01 0.000000e+00
energy 2.600268e-02 1.051887e-04 2.600268e-04
displacement 2.415224e+00 4.243625e-02 1.945777e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
5
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 1.372753e-01 0.000000e+00
energy 2.600268e-02 2.243298e-05 2.600268e-04
displacement 2.415224e+00 5.655989e-02 2.016627e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
6
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 4.153691e-03 0.000000e+00
energy 2.600268e-02 6.211465e-06 2.600268e-04
displacement 2.415224e+00 9.158309e-03 2.097028e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
7
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 2.560894e-04 0.000000e+00
energy 2.600268e-02 5.140028e-07 2.600268e-04
displacement 2.415224e+00 2.051080e-03 2.135940e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
8
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 3.633383e-07 0.000000e+00
energy 2.600268e-02 2.392205e-08 2.600268e-04
displacement 2.415224e+00 9.677880e-05 2.147264e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
9
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 3.252934e-10 0.000000e+00
energy 2.600268e-02 7.696083e-10 2.600268e-04
displacement 2.415224e+00 3.110283e-06 2.149433e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
10
Nonlinear solution status: time= 0.487605
stiffness updates = 0
right hand side evaluations = 13
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.935436e-03 5.592490e-13 0.000000e+00
energy 2.600268e-02 2.184808e-11 2.600268e-04
displacement 2.415224e+00 8.669001e-08 2.149801e-06
convergence summary
number of iterations : 10
number of reformations : 10
------- converged at time : 0.487605
Data Record #1
===========================================================================
Step = 13
Time = 0.487604562
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(49%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0321166
===== beginning time step 14 : 0.519721 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 314983
1
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.562762
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 2.632534e+01 0.000000e+00
energy 2.060417e-02 7.176063e-03 2.060417e-04
displacement 2.145508e+00 6.794856e-01 6.794856e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
2
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 1.225146e+01 0.000000e+00
energy 2.060417e-02 1.617407e-03 2.060417e-04
displacement 2.145508e+00 1.080418e+00 2.903033e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
3
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 6.516283e+00 0.000000e+00
energy 2.060417e-02 1.153451e-03 2.060417e-04
displacement 2.145508e+00 4.410408e-02 2.887797e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
4
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 2.303248e+00 0.000000e+00
energy 2.060417e-02 3.383286e-04 2.060417e-04
displacement 2.145508e+00 4.015758e-01 3.043345e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
5
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 5.198466e-01 0.000000e+00
energy 2.060417e-02 3.103412e-05 2.060417e-04
displacement 2.145508e+00 7.188659e-02 3.227157e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
6
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 4.790416e-03 0.000000e+00
energy 2.060417e-02 9.086751e-06 2.060417e-04
displacement 2.145508e+00 1.692134e-02 3.407742e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
7
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 1.100892e-03 0.000000e+00
energy 2.060417e-02 9.309315e-07 2.060417e-04
displacement 2.145508e+00 5.049656e-03 3.507726e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
8
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 9.806766e-07 0.000000e+00
energy 2.060417e-02 6.600246e-08 2.060417e-04
displacement 2.145508e+00 2.558997e-04 3.536674e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
9
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 1.527133e-09 0.000000e+00
energy 2.060417e-02 2.765888e-09 2.060417e-04
displacement 2.145508e+00 1.084767e-05 3.542718e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
10
Nonlinear solution status: time= 0.519721
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.014944e-03 3.197152e-12 0.000000e+00
energy 2.060417e-02 1.026038e-10 2.060417e-04
displacement 2.145508e+00 3.848279e-07 3.543867e-06
convergence summary
number of iterations : 10
number of reformations : 10
------- converged at time : 0.519721
Data Record #1
===========================================================================
Step = 14
Time = 0.519721148
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(52%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0336933
===== beginning time step 15 : 0.553414 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315025
1
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.538892
convergence norms : INITIAL CURRENT REQUIRED
residual 4.954180e-03 2.989655e+01 0.000000e+00
energy 2.662707e-02 5.345992e-03 2.662707e-04
displacement 2.090303e+00 6.070328e-01 6.070328e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
2
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.954180e-03 3.863263e+00 0.000000e+00
energy 2.662707e-02 1.587093e-03 2.662707e-04
displacement 2.090303e+00 7.020849e-01 2.282359e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
3
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 3
step from line search = 0.500000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.954180e-03 2.332716e+03 0.000000e+00
energy 2.662707e-02 4.353506e-01 2.662707e-04
displacement 2.090303e+00 4.296467e+00 8.220209e-06
*************************************************************************
* WARNING *
* Problem is diverging. Stiffness matrix will now be reformed *
*************************************************************************
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
4
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 2.165368e+00 0.000000e+00
energy 4.353506e-01 5.298220e-03 4.353506e-03
displacement 2.090303e+00 7.120363e-02 7.852230e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
5
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 5
step from line search = 0.074370
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 1.040218e+00 0.000000e+00
energy 4.353506e-01 1.483403e-03 4.353506e-03
displacement 2.090303e+00 1.089671e-01 6.345105e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
6
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 6
step from line search = 0.064557
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 5.537165e-01 0.000000e+00
energy 4.353506e-01 3.995137e-04 4.353506e-03
displacement 2.090303e+00 1.056795e-02 6.011094e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
7
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 7
step from line search = 0.076339
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 2.897432e+00 0.000000e+00
energy 4.353506e-01 7.993131e-04 4.353506e-03
displacement 2.090303e+00 3.695150e-02 5.338061e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
8
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 18
stiffness matrix reformations = 8
step from line search = 0.407420
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 7.367054e+00 0.000000e+00
energy 4.353506e-01 1.313239e-03 4.353506e-03
displacement 2.090303e+00 1.601782e-01 4.089247e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
9
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 19
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 4.226683e+00 0.000000e+00
energy 4.353506e-01 7.465408e-04 4.353506e-03
displacement 2.090303e+00 1.208120e-01 3.414887e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
10
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 20
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 2.954710e+00 0.000000e+00
energy 4.353506e-01 3.911235e-04 4.353506e-03
displacement 2.090303e+00 3.793553e-01 2.451031e-06
Reforming stiffness matrix: reformation #11
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
11
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 21
stiffness matrix reformations = 11
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 1.734936e+00 0.000000e+00
energy 4.353506e-01 1.858252e-04 4.353506e-03
displacement 2.090303e+00 8.477652e-02 2.371495e-06
Reforming stiffness matrix: reformation #12
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
12
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 22
stiffness matrix reformations = 12
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 2.746042e-01 0.000000e+00
energy 4.353506e-01 1.193206e-04 4.353506e-03
displacement 2.090303e+00 9.800089e-02 2.371167e-06
Reforming stiffness matrix: reformation #13
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
13
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 23
stiffness matrix reformations = 13
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 9.637631e-02 0.000000e+00
energy 4.353506e-01 3.471953e-05 4.353506e-03
displacement 2.090303e+00 3.476114e-02 2.515441e-06
Reforming stiffness matrix: reformation #14
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
14
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 24
stiffness matrix reformations = 14
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 4.474477e-03 0.000000e+00
energy 4.353506e-01 7.290253e-06 4.353506e-03
displacement 2.090303e+00 8.380656e-03 2.608796e-06
Reforming stiffness matrix: reformation #15
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
15
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 25
stiffness matrix reformations = 15
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 9.976113e-05 0.000000e+00
energy 4.353506e-01 8.020517e-07 4.353506e-03
displacement 2.090303e+00 1.099217e-03 2.656661e-06
Reforming stiffness matrix: reformation #16
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
16
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 26
stiffness matrix reformations = 16
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 3.352934e-07 0.000000e+00
energy 4.353506e-01 5.107396e-08 4.353506e-03
displacement 2.090303e+00 5.332874e-05 2.672339e-06
Reforming stiffness matrix: reformation #17
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
17
Nonlinear solution status: time= 0.553414
stiffness updates = 0
right hand side evaluations = 27
stiffness matrix reformations = 17
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.332716e+03 5.789340e-10 0.000000e+00
energy 4.353506e-01 1.907011e-09 4.353506e-03
displacement 2.090303e+00 1.845587e-06 2.675670e-06
convergence summary
number of iterations : 17
number of reformations : 17
------- converged at time : 0.553414
Data Record #1
===========================================================================
Step = 15
Time = 0.553414418
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(55%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: decreasing time step, dt = 0.0316736
===== beginning time step 16 : 0.585088 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
1
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.547377
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 3.066111e+01 0.000000e+00
energy 2.902659e-02 3.423075e-03 2.902659e-04
displacement 2.260565e+00 6.773146e-01 6.773146e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
2
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 4.550582e+00 0.000000e+00
energy 2.902659e-02 3.671887e-03 2.902659e-04
displacement 2.260565e+00 7.105383e-01 2.122895e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
3
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.140520
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 2.660044e+00 0.000000e+00
energy 2.902659e-02 2.122573e-04 2.902659e-04
displacement 2.260565e+00 1.022369e-02 2.255535e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
4
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 4
step from line search = 0.191143
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 1.554349e+00 0.000000e+00
energy 2.902659e-02 3.186499e-04 2.902659e-04
displacement 2.260565e+00 5.231311e-03 2.334862e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
5
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 5
step from line search = 0.094679
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 1.563118e+00 0.000000e+00
energy 2.902659e-02 2.538608e-04 2.902659e-04
displacement 2.260565e+00 2.112381e-02 2.206305e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
6
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 6.908647e+00 0.000000e+00
energy 2.902659e-02 9.764677e-04 2.902659e-04
displacement 2.260565e+00 1.216787e-01 2.259080e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
7
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 4.790551e-02 0.000000e+00
energy 2.902659e-02 2.860975e-05 2.902659e-04
displacement 2.260565e+00 8.826583e-03 2.397990e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
8
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 13
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 5.036569e-02 0.000000e+00
energy 2.902659e-02 1.262977e-06 2.902659e-04
displacement 2.260565e+00 4.003042e-03 2.501196e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
9
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 14
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 2.406444e-05 0.000000e+00
energy 2.902659e-02 3.692947e-07 2.902659e-04
displacement 2.260565e+00 4.558575e-04 2.532264e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
10
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 15
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 4.375217e-07 0.000000e+00
energy 2.902659e-02 1.317882e-08 2.902659e-04
displacement 2.260565e+00 5.407728e-05 2.541565e-06
Reforming stiffness matrix: reformation #11
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
11
Nonlinear solution status: time= 0.585088
stiffness updates = 0
right hand side evaluations = 16
stiffness matrix reformations = 11
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 5.258059e-03 8.044741e-11 0.000000e+00
energy 2.902659e-02 4.597668e-10 2.902659e-04
displacement 2.260565e+00 1.265280e-06 2.543604e-06
convergence summary
number of iterations : 11
number of reformations : 11
------- converged at time : 0.585088
Data Record #1
===========================================================================
Step = 16
Time = 0.585087998
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(59%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0330703
===== beginning time step 17 : 0.618158 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315037
1
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.560634
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 2.374575e+01 0.000000e+00
energy 2.070754e-02 4.294814e-03 2.070754e-04
displacement 1.637495e+00 5.146821e-01 5.146821e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315079
2
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 3.112591e+00 0.000000e+00
energy 2.070754e-02 4.611778e-04 2.070754e-04
displacement 1.637495e+00 6.421125e-01 1.882161e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315061
3
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.446836
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 2.874035e+00 0.000000e+00
energy 2.070754e-02 2.230462e-04 2.070754e-04
displacement 1.637495e+00 6.763674e-02 1.775265e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315073
4
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 4.126414e-01 0.000000e+00
energy 2.070754e-02 1.188482e-04 2.070754e-04
displacement 1.637495e+00 2.837292e-02 1.995918e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315061
5
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 1.407048e-01 0.000000e+00
energy 2.070754e-02 5.182441e-05 2.070754e-04
displacement 1.637495e+00 6.336593e-02 2.255907e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315061
6
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 2.696642e-02 0.000000e+00
energy 2.070754e-02 1.434133e-05 2.070754e-04
displacement 1.637495e+00 2.186579e-02 2.445442e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315061
7
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 1.237627e-03 0.000000e+00
energy 2.070754e-02 2.376078e-06 2.070754e-04
displacement 1.637495e+00 5.678509e-03 2.550626e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315073
8
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 1.446784e-05 0.000000e+00
energy 2.070754e-02 1.897125e-07 2.070754e-04
displacement 1.637495e+00 7.197058e-04 2.590419e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315073
9
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 2.283772e-08 0.000000e+00
energy 2.070754e-02 1.289855e-08 2.070754e-04
displacement 1.637495e+00 4.775766e-05 2.601900e-06
Reforming stiffness matrix: reformation #10
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315073
10
Nonlinear solution status: time= 0.618158
stiffness updates = 0
right hand side evaluations = 13
stiffness matrix reformations = 10
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.471810e-03 6.085389e-11 0.000000e+00
energy 2.070754e-02 7.264506e-10 2.070754e-04
displacement 1.637495e+00 2.572094e-06 2.604605e-06
convergence summary
number of iterations : 10
number of reformations : 10
------- converged at time : 0.618158
Data Record #1
===========================================================================
Step = 17
Time = 0.618158323
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(62%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0344563
===== beginning time step 18 : 0.652615 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315073
1
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.569164
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 1.364057e+01 0.000000e+00
energy 1.262259e-02 1.767199e-03 1.262259e-04
displacement 9.855984e-01 3.192828e-01 3.192828e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
2
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 1.117779e+01 0.000000e+00
energy 1.262259e-02 7.991999e-04 1.262259e-04
displacement 9.855984e-01 9.293450e-01 1.508213e-06
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
3
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 8.860053e-01 0.000000e+00
energy 1.262259e-02 1.482920e-04 1.262259e-04
displacement 9.855984e-01 1.654787e-02 1.674581e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
4
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 7.390249e-01 0.000000e+00
energy 1.262259e-02 3.308214e-05 1.262259e-04
displacement 9.855984e-01 1.660326e-01 2.375292e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
5
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 7.155413e-02 0.000000e+00
energy 1.262259e-02 2.967341e-05 1.262259e-04
displacement 9.855984e-01 2.353293e-02 2.689142e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
6
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 8.604103e-03 0.000000e+00
energy 1.262259e-02 6.797623e-06 1.262259e-04
displacement 9.855984e-01 1.288908e-02 2.955399e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
7
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 2.166766e-04 0.000000e+00
energy 1.262259e-02 6.657745e-07 1.262259e-04
displacement 9.855984e-01 1.610819e-03 3.056635e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
8
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 4.277612e-07 0.000000e+00
energy 1.262259e-02 2.870023e-08 1.262259e-04
displacement 9.855984e-01 9.045034e-05 3.083780e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
9
Nonlinear solution status: time= 0.652615
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.938369e-03 2.460596e-10 0.000000e+00
energy 1.262259e-02 8.619571e-10 1.262259e-04
displacement 9.855984e-01 2.356794e-06 3.088541e-06
convergence summary
number of iterations : 9
number of reformations : 9
------- converged at time : 0.652615
Data Record #1
===========================================================================
Step = 18
Time = 0.652614583
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(65%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.035565
===== beginning time step 19 : 0.68818 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
1
Nonlinear solution status: time= 0.68818
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.590768
convergence norms : INITIAL CURRENT REQUIRED
residual 1.577312e-03 1.111270e+01 0.000000e+00
energy 9.936452e-03 1.265458e-03 9.936452e-05
displacement 7.079639e-01 2.470838e-01 2.470838e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
2
Nonlinear solution status: time= 0.68818
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.577312e-03 1.841415e+00 0.000000e+00
energy 9.936452e-03 1.798135e-04 9.936452e-05
displacement 7.079639e-01 4.543468e-01 9.900283e-07
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
3
Nonlinear solution status: time= 0.68818
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.577312e-03 2.443210e+00 0.000000e+00
energy 9.936452e-03 2.862138e-04 9.936452e-05
displacement 7.079639e-01 5.441932e-02 1.119598e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
4
Nonlinear solution status: time= 0.68818
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.577312e-03 2.525205e-02 0.000000e+00
energy 9.936452e-03 2.400164e-05 9.936452e-05
displacement 7.079639e-01 2.001485e-02 1.290679e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
5
Nonlinear solution status: time= 0.68818
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.577312e-03 3.151520e-03 0.000000e+00
energy 9.936452e-03 2.797504e-06 9.936452e-05
displacement 7.079639e-01 5.028419e-03 1.375990e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
6
Nonlinear solution status: time= 0.68818
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.577312e-03 1.150773e-05 0.000000e+00
energy 9.936452e-03 1.545063e-07 9.936452e-05
displacement 7.079639e-01 2.693754e-04 1.401932e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
7
Nonlinear solution status: time= 0.68818
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.577312e-03 7.114905e-09 0.000000e+00
energy 9.936452e-03 3.208020e-09 9.936452e-05
displacement 7.079639e-01 6.592958e-06 1.406478e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
8
Nonlinear solution status: time= 0.68818
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.577312e-03 1.355198e-12 0.000000e+00
energy 9.936452e-03 3.760004e-11 9.936452e-05
displacement 7.079639e-01 7.688317e-08 1.407013e-06
convergence summary
number of iterations : 8
number of reformations : 8
------- converged at time : 0.68818
Data Record #1
===========================================================================
Step = 19
Time = 0.68817959
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(69%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.036452
===== beginning time step 20 : 0.724632 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
1
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.574825
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 1.031941e+01 0.000000e+00
energy 1.026895e-02 1.307935e-03 1.026895e-04
displacement 8.059678e-01 2.663109e-01 2.663109e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315121
2
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 3.659292e+00 0.000000e+00
energy 1.026895e-02 3.233844e-04 1.026895e-04
displacement 8.059678e-01 3.456098e-01 9.591622e-07
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
3
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 3
step from line search = 0.132818
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 3.008269e+00 0.000000e+00
energy 1.026895e-02 1.230595e-04 1.026895e-04
displacement 8.059678e-01 1.186431e-02 9.392214e-07
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
4
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 5.909542e-01 0.000000e+00
energy 1.026895e-02 1.608727e-05 1.026895e-04
displacement 8.059678e-01 3.251502e-02 1.095735e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
5
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 2.849773e-02 0.000000e+00
energy 1.026895e-02 1.539988e-05 1.026895e-04
displacement 8.059678e-01 1.669459e-02 1.239868e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
6
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 4.714121e-03 0.000000e+00
energy 1.026895e-02 1.278149e-06 1.026895e-04
displacement 8.059678e-01 5.228745e-03 1.290203e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
7
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 2.734011e-06 0.000000e+00
energy 1.026895e-02 7.392679e-08 1.026895e-04
displacement 8.059678e-01 2.263215e-04 1.304633e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
8
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 11
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 1.369618e-09 0.000000e+00
energy 1.026895e-02 1.144037e-09 1.026895e-04
displacement 8.059678e-01 6.029546e-06 1.306755e-06
Reforming stiffness matrix: reformation #9
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
9
Nonlinear solution status: time= 0.724632
stiffness updates = 0
right hand side evaluations = 12
stiffness matrix reformations = 9
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.372277e-03 1.843050e-13 0.000000e+00
energy 1.026895e-02 1.531228e-11 1.026895e-04
displacement 8.059678e-01 5.683970e-08 1.306972e-06
convergence summary
number of iterations : 9
number of reformations : 9
------- converged at time : 0.724632
Data Record #1
===========================================================================
Step = 20
Time = 0.724631597
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(72%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0371616
===== beginning time step 21 : 0.761793 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315091
1
Nonlinear solution status: time= 0.761793
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.605394
convergence norms : INITIAL CURRENT REQUIRED
residual 6.729605e-04 5.491779e+00 0.000000e+00
energy 5.739610e-03 5.092835e-04 5.739610e-05
displacement 4.916434e-01 1.801880e-01 1.801880e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
2
Nonlinear solution status: time= 0.761793
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.729605e-04 1.016085e+01 0.000000e+00
energy 5.739610e-03 1.226383e-03 5.739610e-05
displacement 4.916434e-01 7.184710e-01 9.407260e-07
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
3
Nonlinear solution status: time= 0.761793
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.729605e-04 1.026827e-01 0.000000e+00
energy 5.739610e-03 1.022185e-04 5.739610e-05
displacement 4.916434e-01 1.180605e-02 1.062046e-06
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
4
Nonlinear solution status: time= 0.761793
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.729605e-04 2.304177e-01 0.000000e+00
energy 5.739610e-03 7.009093e-07 5.739610e-05
displacement 4.916434e-01 3.562110e-02 1.359849e-06
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
5
Nonlinear solution status: time= 0.761793
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.729605e-04 1.575974e-03 0.000000e+00
energy 5.739610e-03 5.514878e-06 5.739610e-05
displacement 4.916434e-01 3.275975e-03 1.454529e-06
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
6
Nonlinear solution status: time= 0.761793
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.729605e-04 2.393171e-04 0.000000e+00
energy 5.739610e-03 3.755541e-07 5.739610e-05
displacement 4.916434e-01 1.155162e-03 1.510973e-06
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
7
Nonlinear solution status: time= 0.761793
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.729605e-04 7.820450e-08 0.000000e+00
energy 5.739610e-03 1.158217e-08 5.739610e-05
displacement 4.916434e-01 3.725374e-05 1.523523e-06
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
8
Nonlinear solution status: time= 0.761793
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 6.729605e-04 2.159878e-11 0.000000e+00
energy 5.739610e-03 2.131704e-10 5.739610e-05
displacement 4.916434e-01 7.138921e-07 1.525336e-06
convergence summary
number of iterations : 8
number of reformations : 8
------- converged at time : 0.761793
Data Record #1
===========================================================================
Step = 21
Time = 0.761793201
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(76%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0377293
===== beginning time step 22 : 0.799522 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
1
Nonlinear solution status: time= 0.799522
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.619088
convergence norms : INITIAL CURRENT REQUIRED
residual 3.446212e-04 1.925308e+00 0.000000e+00
energy 3.065880e-03 2.109609e-04 3.065880e-05
displacement 3.291425e-01 1.261505e-01 1.261505e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315145
2
Nonlinear solution status: time= 0.799522
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.446212e-04 4.565041e+00 0.000000e+00
energy 3.065880e-03 3.975534e-04 3.065880e-05
displacement 3.291425e-01 4.702095e-01 3.633815e-07
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
3
Nonlinear solution status: time= 0.799522
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.446212e-04 3.418568e-02 0.000000e+00
energy 3.065880e-03 4.209789e-05 3.065880e-05
displacement 3.291425e-01 6.681891e-03 4.382643e-07
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
4
Nonlinear solution status: time= 0.799522
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.446212e-04 2.210502e-02 0.000000e+00
energy 3.065880e-03 9.275677e-07 3.065880e-05
displacement 3.291425e-01 1.066690e-02 5.328483e-07
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
5
Nonlinear solution status: time= 0.799522
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.446212e-04 4.376372e-06 0.000000e+00
energy 3.065880e-03 1.474590e-07 3.065880e-05
displacement 3.291425e-01 2.198550e-04 5.505191e-07
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
6
Nonlinear solution status: time= 0.799522
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.446212e-04 1.114756e-08 0.000000e+00
energy 3.065880e-03 2.223269e-09 3.065880e-05
displacement 3.291425e-01 9.153464e-06 5.537771e-07
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
7
Nonlinear solution status: time= 0.799522
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 3.446212e-04 6.509393e-13 0.000000e+00
energy 3.065880e-03 2.592324e-11 3.065880e-05
displacement 3.291425e-01 8.218816e-08 5.541457e-07
convergence summary
number of iterations : 7
number of reformations : 7
------- converged at time : 0.799522
Data Record #1
===========================================================================
Step = 22
Time = 0.799522485
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(80%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0381834
===== beginning time step 23 : 0.837706 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
1
Nonlinear solution status: time= 0.837706
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.606185
convergence norms : INITIAL CURRENT REQUIRED
residual 2.257341e-04 1.149652e+00 0.000000e+00
energy 2.311147e-03 3.077682e-04 2.311147e-05
displacement 3.047026e-01 1.119661e-01 1.119661e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
2
Nonlinear solution status: time= 0.837706
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.257341e-04 2.370116e+00 0.000000e+00
energy 2.311147e-03 4.590061e-05 2.311147e-05
displacement 3.047026e-01 2.433070e-01 1.911420e-07
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
3
Nonlinear solution status: time= 0.837706
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.257341e-04 2.041365e-02 0.000000e+00
energy 2.311147e-03 2.978443e-05 2.311147e-05
displacement 3.047026e-01 7.819194e-03 2.436522e-07
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
4
Nonlinear solution status: time= 0.837706
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.257341e-04 5.199001e-03 0.000000e+00
energy 2.311147e-03 1.424628e-06 2.311147e-05
displacement 3.047026e-01 3.008754e-03 2.790988e-07
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
5
Nonlinear solution status: time= 0.837706
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.257341e-04 7.297330e-07 0.000000e+00
energy 2.311147e-03 4.030564e-08 2.311147e-05
displacement 3.047026e-01 6.304953e-05 2.852801e-07
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
6
Nonlinear solution status: time= 0.837706
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.257341e-04 2.795061e-10 0.000000e+00
energy 2.311147e-03 3.778994e-10 2.311147e-05
displacement 3.047026e-01 1.051075e-06 2.861049e-07
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
7
Nonlinear solution status: time= 0.837706
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 2.257341e-04 2.526369e-14 0.000000e+00
energy 2.311147e-03 2.838372e-12 2.311147e-05
displacement 3.047026e-01 8.583658e-09 2.861832e-07
convergence summary
number of iterations : 7
number of reformations : 7
------- converged at time : 0.837706
Data Record #1
===========================================================================
Step = 23
Time = 0.837705913
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(84%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0385467
===== beginning time step 24 : 0.876253 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315133
1
Nonlinear solution status: time= 0.876253
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.553991
convergence norms : INITIAL CURRENT REQUIRED
residual 1.451642e-04 6.533264e-01 0.000000e+00
energy 1.687031e-03 3.977698e-04 1.687031e-05
displacement 3.212318e-01 9.858792e-02 9.858792e-08
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
2
Nonlinear solution status: time= 0.876253
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.451642e-04 2.371150e+00 0.000000e+00
energy 1.687031e-03 2.123791e-04 1.687031e-05
displacement 3.212318e-01 3.065592e-01 1.340872e-07
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
3
Nonlinear solution status: time= 0.876253
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.451642e-04 9.423983e-03 0.000000e+00
energy 1.687031e-03 1.287393e-05 1.687031e-05
displacement 3.212318e-01 4.970695e-03 1.704347e-07
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
4
Nonlinear solution status: time= 0.876253
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.451642e-04 7.544385e-04 0.000000e+00
energy 1.687031e-03 5.703614e-07 1.687031e-05
displacement 3.212318e-01 9.671897e-04 1.912433e-07
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
5
Nonlinear solution status: time= 0.876253
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.451642e-04 3.719536e-08 0.000000e+00
energy 1.687031e-03 1.182446e-08 1.687031e-05
displacement 3.212318e-01 2.932615e-05 1.952186e-07
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
6
Nonlinear solution status: time= 0.876253
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.451642e-04 1.874999e-11 0.000000e+00
energy 1.687031e-03 2.368903e-10 1.687031e-05
displacement 3.212318e-01 7.134113e-07 1.958726e-07
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
7
Nonlinear solution status: time= 0.876253
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.451642e-04 5.324243e-14 0.000000e+00
energy 1.687031e-03 4.592536e-12 1.687031e-05
displacement 3.212318e-01 1.441361e-08 1.959657e-07
convergence summary
number of iterations : 7
number of reformations : 7
------- converged at time : 0.876253
Data Record #1
===========================================================================
Step = 24
Time = 0.876252654
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(88%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0388374
===== beginning time step 25 : 0.91509 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
1
Nonlinear solution status: time= 0.91509
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.499020
convergence norms : INITIAL CURRENT REQUIRED
residual 1.405541e-04 6.758369e-01 0.000000e+00
energy 2.034191e-03 7.181996e-04 2.034191e-05
displacement 9.263767e-01 2.306871e-01 2.306871e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
2
Nonlinear solution status: time= 0.91509
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.405541e-04 6.536125e-01 0.000000e+00
energy 2.034191e-03 8.578294e-05 2.034191e-05
displacement 9.263767e-01 3.079466e-01 1.809985e-08
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
3
Nonlinear solution status: time= 0.91509
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.405541e-04 1.199548e-02 0.000000e+00
energy 2.034191e-03 2.168782e-05 2.034191e-05
displacement 9.263767e-01 8.388264e-03 3.603553e-08
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
4
Nonlinear solution status: time= 0.91509
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.405541e-04 1.347311e-03 0.000000e+00
energy 2.034191e-03 1.200073e-06 2.034191e-05
displacement 9.263767e-01 1.419105e-03 4.837087e-08
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
5
Nonlinear solution status: time= 0.91509
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.405541e-04 3.028539e-07 0.000000e+00
energy 2.034191e-03 1.957261e-08 2.034191e-05
displacement 9.263767e-01 4.637179e-05 5.095252e-08
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
6
Nonlinear solution status: time= 0.91509
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.405541e-04 3.277719e-11 0.000000e+00
energy 2.034191e-03 1.999471e-10 2.034191e-05
displacement 9.263767e-01 7.956060e-07 5.130657e-08
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
7
Nonlinear solution status: time= 0.91509
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.405541e-04 1.534655e-14 0.000000e+00
energy 2.034191e-03 2.012152e-12 2.034191e-05
displacement 9.263767e-01 8.480807e-09 5.134303e-08
convergence summary
number of iterations : 7
number of reformations : 7
------- converged at time : 0.91509
Data Record #1
===========================================================================
Step = 25
Time = 0.915090048
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(92%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0390699
===== beginning time step 26 : 0.95416 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
1
Nonlinear solution status: time= 0.95416
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.503837
convergence norms : INITIAL CURRENT REQUIRED
residual 1.393630e-04 7.262306e-01 0.000000e+00
energy 2.060397e-03 6.504072e-04 2.060397e-05
displacement 9.077755e-01 2.304408e-01 2.304408e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
2
Nonlinear solution status: time= 0.95416
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.393630e-04 1.766826e-01 0.000000e+00
energy 2.060397e-03 9.313616e-05 2.060397e-05
displacement 9.077755e-01 2.012687e-01 3.313014e-09
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
3
Nonlinear solution status: time= 0.95416
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.393630e-04 9.433462e-03 0.000000e+00
energy 2.060397e-03 1.030833e-05 2.060397e-05
displacement 9.077755e-01 7.159139e-03 3.201138e-09
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
4
Nonlinear solution status: time= 0.95416
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.393630e-04 5.352371e-05 0.000000e+00
energy 2.060397e-03 3.612435e-07 2.060397e-05
displacement 9.077755e-01 5.254857e-04 5.665027e-09
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
5
Nonlinear solution status: time= 0.95416
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.393630e-04 1.431614e-08 0.000000e+00
energy 2.060397e-03 3.882686e-09 2.060397e-05
displacement 9.077755e-01 1.453353e-05 6.188322e-09
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
6
Nonlinear solution status: time= 0.95416
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.393630e-04 7.543506e-13 0.000000e+00
energy 2.060397e-03 3.753617e-11 2.060397e-05
displacement 9.077755e-01 1.646003e-07 6.246306e-09
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
7
Nonlinear solution status: time= 0.95416
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.393630e-04 1.911877e-15 0.000000e+00
energy 2.060397e-03 2.986532e-13 2.060397e-05
displacement 9.077755e-01 1.309404e-09 6.251476e-09
convergence summary
number of iterations : 7
number of reformations : 7
------- converged at time : 0.95416
Data Record #1
===========================================================================
Step = 26
Time = 0.954159962
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(95%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0392559
===== beginning time step 27 : 0.993416 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
1
Nonlinear solution status: time= 0.993416
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.505367
convergence norms : INITIAL CURRENT REQUIRED
residual 1.412206e-04 7.580302e-01 0.000000e+00
energy 2.095309e-03 6.262617e-04 2.095309e-05
displacement 9.077166e-01 2.318268e-01 2.318268e-07
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
2
Nonlinear solution status: time= 0.993416
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.412206e-04 1.153972e-01 0.000000e+00
energy 2.095309e-03 8.444773e-05 2.095309e-05
displacement 9.077166e-01 1.531805e-01 9.507366e-09
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
3
Nonlinear solution status: time= 0.993416
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.412206e-04 7.729946e-03 0.000000e+00
energy 2.095309e-03 7.650415e-06 2.095309e-05
displacement 9.077166e-01 6.079282e-03 3.889439e-10
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
4
Nonlinear solution status: time= 0.993416
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.412206e-04 1.759745e-05 0.000000e+00
energy 2.095309e-03 2.199806e-07 2.095309e-05
displacement 9.077166e-01 3.203863e-04 1.257878e-11
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
5
Nonlinear solution status: time= 0.993416
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.412206e-04 4.387292e-09 0.000000e+00
energy 2.095309e-03 2.244873e-09 2.095309e-05
displacement 9.077166e-01 7.974251e-06 1.204072e-11
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
6
Nonlinear solution status: time= 0.993416
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.412206e-04 3.024428e-13 0.000000e+00
energy 2.095309e-03 2.163092e-11 2.095309e-05
displacement 9.077166e-01 8.976274e-08 1.311526e-11
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
7
Nonlinear solution status: time= 0.993416
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.412206e-04 1.108822e-15 0.000000e+00
energy 2.095309e-03 1.741324e-13 2.095309e-05
displacement 9.077166e-01 7.221748e-10 1.322140e-11
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
8
Nonlinear solution status: time= 0.993416
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 1.412206e-04 8.645307e-18 0.000000e+00
energy 2.095309e-03 1.370473e-15 2.095309e-05
displacement 9.077166e-01 5.678214e-12 1.323088e-11
convergence summary
number of iterations : 8
number of reformations : 8
------- converged at time : 0.993416
Data Record #1
===========================================================================
Step = 27
Time = 0.993415894
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(99%) tempModel.feb - FEBio 4.1.0
AUTO STEPPER: increasing time step, dt = 0.0394047
MUST POINT CONTROLLER: adjusting time step. dt = 0.00658411
===== beginning time step 28 : 1 =====
Reforming stiffness matrix: reformation #1
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
1
Nonlinear solution status: time= 1
stiffness updates = 0
right hand side evaluations = 3
stiffness matrix reformations = 1
step from line search = 0.630060
convergence norms : INITIAL CURRENT REQUIRED
residual 4.266843e-06 1.687311e-03 0.000000e+00
energy 6.437406e-05 2.347021e-05 6.437406e-07
displacement 2.676759e-02 1.062609e-02 1.062609e-08
Reforming stiffness matrix: reformation #2
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
2
Nonlinear solution status: time= 1
stiffness updates = 0
right hand side evaluations = 4
stiffness matrix reformations = 2
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.266843e-06 1.031982e-03 0.000000e+00
energy 6.437406e-05 2.104634e-06 6.437406e-07
displacement 2.676759e-02 8.678732e-03 9.892171e-11
Reforming stiffness matrix: reformation #3
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
3
Nonlinear solution status: time= 1
stiffness updates = 0
right hand side evaluations = 5
stiffness matrix reformations = 3
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.266843e-06 2.006082e-07 0.000000e+00
energy 6.437406e-05 2.458944e-08 6.437406e-07
displacement 2.676759e-02 8.019419e-05 9.819203e-13
Reforming stiffness matrix: reformation #4
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
4
Nonlinear solution status: time= 1
stiffness updates = 0
right hand side evaluations = 6
stiffness matrix reformations = 4
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.266843e-06 2.216502e-11 0.000000e+00
energy 6.437406e-05 2.091587e-10 6.437406e-07
displacement 2.676759e-02 8.099874e-07 8.289959e-15
Reforming stiffness matrix: reformation #5
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
5
Nonlinear solution status: time= 1
stiffness updates = 0
right hand side evaluations = 7
stiffness matrix reformations = 5
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.266843e-06 1.149555e-14 0.000000e+00
energy 6.437406e-05 1.670150e-12 6.437406e-07
displacement 2.676759e-02 6.919902e-09 6.183101e-17
Reforming stiffness matrix: reformation #6
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
6
Nonlinear solution status: time= 1
stiffness updates = 0
right hand side evaluations = 8
stiffness matrix reformations = 6
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.266843e-06 8.285381e-17 0.000000e+00
energy 6.437406e-05 1.313586e-14 6.437406e-07
displacement 2.676759e-02 5.442488e-11 2.361454e-19
Reforming stiffness matrix: reformation #7
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
7
Nonlinear solution status: time= 1
stiffness updates = 0
right hand side evaluations = 9
stiffness matrix reformations = 7
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.266843e-06 6.486957e-19 0.000000e+00
energy 6.437406e-05 1.032822e-16 6.437406e-07
displacement 2.676759e-02 4.279169e-13 2.829313e-20
Reforming stiffness matrix: reformation #8
===== reforming stiffness matrix:
Nr of equations ........................... : 4351
Nr of nonzeroes in stiffness matrix ....... : 315163
8
Nonlinear solution status: time= 1
stiffness updates = 0
right hand side evaluations = 10
stiffness matrix reformations = 8
step from line search = 1.000000
convergence norms : INITIAL CURRENT REQUIRED
residual 4.266843e-06 5.292859e-21 0.000000e+00
energy 6.437406e-05 8.120748e-19 6.437406e-07
displacement 2.676759e-02 3.364580e-15 5.117125e-20
*************************************************************************
* WARNING *
* No force acting on the system. *
*************************************************************************
convergence summary
number of iterations : 8
number of reformations : 8
------- converged at time : 1
Data Record #1
===========================================================================
Step = 28
Time = 1
Data = ux;uy;uz
File = /mnt/data/MATLAB/GIBBON/data/temp/tempModel_disp_out.txt
]0;(100%) tempModel.feb - FEBio 4.1.0
N O N L I N E A R I T E R A T I O N I N F O R M A T I O N
Number of time steps completed .................... : 28
Total number of equilibrium iterations ............ : 237
Average number of equilibrium iterations .......... : 8.46429
Total number of right hand evaluations ............ : 342
Total number of stiffness reformations ............ : 237
L I N E A R S O L V E R S T A T S
Total calls to linear solver ........ : 286
Avg iterations per solve ............ : 1
Time in linear solver: 0:00:20
Elapsed time : 0:00:33
N O R M A L T E R M I N A T I O N
]0;(100%) tempModel.feb - FEBio 4.1.0 * Log file found. 20-Apr-2023 18:02:21
# Parsing log file... 20-Apr-2023 18:02:21
number of iterations : 1 20-Apr-2023 18:02:22
number of reformations : 1 20-Apr-2023 18:02:22
------- converged at time : 0.04 20-Apr-2023 18:02:22
number of iterations : 1 20-Apr-2023 18:02:22
number of reformations : 1 20-Apr-2023 18:02:22
------- converged at time : 0.08 20-Apr-2023 18:02:22
number of iterations : 1 20-Apr-2023 18:02:22
number of reformations : 1 20-Apr-2023 18:02:22
------- converged at time : 0.12 20-Apr-2023 18:02:22
number of iterations : 1 20-Apr-2023 18:02:22
number of reformations : 1 20-Apr-2023 18:02:22
------- converged at time : 0.16 20-Apr-2023 18:02:22
number of iterations : 8 20-Apr-2023 18:02:22
number of reformations : 8 20-Apr-2023 18:02:22
------- converged at time : 0.2 20-Apr-2023 18:02:22
number of iterations : 11 20-Apr-2023 18:02:22
number of reformations : 11 20-Apr-2023 18:02:22
------- converged at time : 0.238462 20-Apr-2023 18:02:22
number of iterations : 13 20-Apr-2023 18:02:22
number of reformations : 13 20-Apr-2023 18:02:22
------- converged at time : 0.275692 20-Apr-2023 18:02:22
number of iterations : 10 20-Apr-2023 18:02:22
number of reformations : 10 20-Apr-2023 18:02:22
------- converged at time : 0.313128 20-Apr-2023 18:02:22
number of iterations : 12 20-Apr-2023 18:02:22
number of reformations : 12 20-Apr-2023 18:02:22
------- converged at time : 0.351076 20-Apr-2023 18:02:22
number of iterations : 12 20-Apr-2023 18:02:22
number of reformations : 12 20-Apr-2023 18:02:22
------- converged at time : 0.389267 20-Apr-2023 18:02:22
number of iterations : 10 20-Apr-2023 18:02:22
number of reformations : 10 20-Apr-2023 18:02:22
------- converged at time : 0.427672 20-Apr-2023 18:02:22
number of iterations : 14 20-Apr-2023 18:02:22
number of reformations : 14 20-Apr-2023 18:02:22
------- converged at time : 0.457459 20-Apr-2023 18:02:22
number of iterations : 10 20-Apr-2023 18:02:22
number of reformations : 10 20-Apr-2023 18:02:22
------- converged at time : 0.487605 20-Apr-2023 18:02:22
number of iterations : 10 20-Apr-2023 18:02:22
number of reformations : 10 20-Apr-2023 18:02:22
------- converged at time : 0.519721 20-Apr-2023 18:02:22
number of iterations : 17 20-Apr-2023 18:02:22
number of reformations : 17 20-Apr-2023 18:02:22
------- converged at time : 0.553414 20-Apr-2023 18:02:22
number of iterations : 11 20-Apr-2023 18:02:22
number of reformations : 11 20-Apr-2023 18:02:22
------- converged at time : 0.585088 20-Apr-2023 18:02:22
number of iterations : 10 20-Apr-2023 18:02:22
number of reformations : 10 20-Apr-2023 18:02:22
------- converged at time : 0.618158 20-Apr-2023 18:02:22
number of iterations : 9 20-Apr-2023 18:02:22
number of reformations : 9 20-Apr-2023 18:02:22
------- converged at time : 0.652615 20-Apr-2023 18:02:22
number of iterations : 8 20-Apr-2023 18:02:22
number of reformations : 8 20-Apr-2023 18:02:22
------- converged at time : 0.68818 20-Apr-2023 18:02:22
number of iterations : 9 20-Apr-2023 18:02:22
number of reformations : 9 20-Apr-2023 18:02:22
------- converged at time : 0.724632 20-Apr-2023 18:02:22
number of iterations : 8 20-Apr-2023 18:02:22
number of reformations : 8 20-Apr-2023 18:02:22
------- converged at time : 0.761793 20-Apr-2023 18:02:22
number of iterations : 7 20-Apr-2023 18:02:22
number of reformations : 7 20-Apr-2023 18:02:22
------- converged at time : 0.799522 20-Apr-2023 18:02:22
number of iterations : 7 20-Apr-2023 18:02:22
number of reformations : 7 20-Apr-2023 18:02:22
------- converged at time : 0.837706 20-Apr-2023 18:02:22
number of iterations : 7 20-Apr-2023 18:02:22
number of reformations : 7 20-Apr-2023 18:02:22
------- converged at time : 0.876253 20-Apr-2023 18:02:22
number of iterations : 7 20-Apr-2023 18:02:22
number of reformations : 7 20-Apr-2023 18:02:22
------- converged at time : 0.91509 20-Apr-2023 18:02:22
number of iterations : 7 20-Apr-2023 18:02:22
number of reformations : 7 20-Apr-2023 18:02:22
------- converged at time : 0.95416 20-Apr-2023 18:02:22
number of iterations : 8 20-Apr-2023 18:02:22
number of reformations : 8 20-Apr-2023 18:02:22
------- converged at time : 0.993416 20-Apr-2023 18:02:22
number of iterations : 8 20-Apr-2023 18:02:22
number of reformations : 8 20-Apr-2023 18:02:22
------- converged at time : 1 20-Apr-2023 18:02:22
Elapsed time : 0:00:33 20-Apr-2023 18:02:22
N O R M A L T E R M I N A T I O N
# Done 20-Apr-2023 18:02:22
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
Import FEBio results
if 1%runFlag==1 %i.e. a succesful run
Importing nodal displacements from a log file
dataStruct=importFEBio_logfile(fullfile(savePath,febioLogFileName_disp),0,1);
%Access data
N_disp_mat=dataStruct.data; %Displacement
timeVec=dataStruct.time; %Time
%Create deformed coordinate set
V_DEF=N_disp_mat+repmat(V,[1 1 size(N_disp_mat,3)]);
Plotting the simulated results using anim8 to visualize and animate deformations
DN_magnitude=sqrt(sum(N_disp_mat(:,:,end).^2,2)); %Current displacement magnitude % Create basic view and store graphics handle to initiate animation hf=cFigure; hold on; gtitle([febioFebFileNamePart,': Press play to animate']); hp1=gpatch(Fb_blob_plot,V_DEF(:,:,end),DN_magnitude,'k',1); %Add graphics object to animate hp1.FaceColor='interp'; hp2=gpatch(F_probe_plot,V_DEF(:,:,end),'kw','none',0.5); %Add graphics object to animate % gpatch(Fb_all,V,0.5*ones(1,3),'none',0.25); %A static graphics object axisGeom(gca,fontSize); colormap(gjet(250)); colorbar; caxis([0 max(DN_magnitude(Fb_blob_plot(:)))]); caxis manual; axis(axisLim(V_DEF)); %Set axis limits statically camlight headlight; view(15,8); drawnow; % Set up animation features animStruct.Time=timeVec; %The time vector for qt=1:1:size(N_disp_mat,3) %Loop over time increments DN_magnitude=sqrt(sum(N_disp_mat(:,:,qt).^2,2)); %Current displacement magnitude %Set entries in animation structure animStruct.Handles{qt}=[hp1 hp1 hp2]; %Handles of objects to animate animStruct.Props{qt}={'Vertices','CData','Vertices'}; %Properties of objects to animate animStruct.Set{qt}={V_DEF(:,:,qt),DN_magnitude,V_DEF(:,:,qt)}; %Property values for to set in order to animate end anim8(hf,animStruct); %Initiate animation feature drawnow;

end

GIBBON www.gibboncode.org
Kevin Mattheus Moerman, [email protected]
GIBBON footer text
License: https://github.com/gibbonCode/GIBBON/blob/master/LICENSE
GIBBON: The Geometry and Image-based Bioengineering add-On. A toolbox for image segmentation, image-based modeling, meshing, and finite element analysis.
Copyright (C) 2006-2022 Kevin Mattheus Moerman and the GIBBON contributors
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.